Related Research Articles

In genetics, a fusion gene is a hybrid gene formed from two previously independent genes. It can occur as a result of translocation, interstitial deletion, or chromosomal inversion. Fusion genes have been found to be prevalent in all main types of human neoplasia. The identification of these fusion genes play a prominent role in being a diagnostic and prognostic marker.
Double minutes (DMs) are small fragments of extrachromosomal DNA, which have been observed in a large number of human tumors including breast, lung, ovary, colon, and most notably, neuroblastoma. They are a manifestation of gene amplification as a result of chromothripsis, during the development of tumors, which give the cells selective advantages for growth and survival. This selective advantage is as a result of double minutes frequently harboring amplified oncogenes and genes involved in drug resistance. DMs, like actual chromosomes, are composed of chromatin and replicate in the nucleus of the cell during cell division. Unlike typical chromosomes, they are composed of circular fragments of DNA, up to only a few million base pairs in size, and contain no centromere or telomere. Further to this, they often lack key regulatory elements, allowing genes to be constitutively expressed. The term ecDNA may be used to refer to DMs in a more general manner. The term Double Minute originates from the visualization of these features under microscope; double because the dots were found in pairs, and minute because they were minuscule.

Ewing sarcoma is a type of pediatric cancer that forms in bone or soft tissue. Symptoms may include swelling and pain at the site of the tumor, fever, and a bone fracture. The most common areas where it begins are the legs, pelvis, and chest wall. In about 25% of cases, the cancer has already spread to other parts of the body at the time of diagnosis. Complications may include a pleural effusion or paraplegia.

High-mobility group AT-hook 2, also known as HMGA2, is a protein that, in humans, is encoded by the HMGA2 gene.
RNA-binding protein EWS is a protein that in humans is encoded by the EWSR1 gene on human chromosome 22, specifically 22q12.2. It is one of 3 proteins in the FET protein family.

ERG is an oncogene. ERG is a member of the ETS family of transcription factors. The ERG gene encodes for a protein, also called ERG, that functions as a transcriptional regulator. Genes in the ETS family regulate embryonic development, cell proliferation, differentiation, angiogenesis, inflammation, and apoptosis.

Transmembrane protease, serine 2 is an enzyme that in humans is encoded by the TMPRSS2 gene. It belongs to the TMPRSS family of proteins, whose members are transmembrane proteins which have a serine protease activity. The TMPRSS2 protein is found in high concentration in the cell membranes of epithelial cells of the lung and of the prostate, but also in the heart, liver and gastrointestinal tract.

Zinc finger protein 384 is a protein that in humans is encoded by the ZNF384 gene.
In the field of molecular biology, the ETSfamily is one of the largest families of transcription factors and is unique to animals. There are 28 genes in humans, 27 in the mouse, 10 in Caenorhabditis elegans and 9 in Drosophila. The founding member of this family was identified as a gene transduced by the leukemia virus, E26. The members of the family have been implicated in the development of different tissues as well as cancer progression.
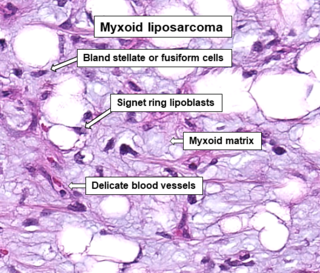
A myxoid liposarcoma is a malignant adipose tissue neoplasm of myxoid appearance histologically.

Solute carrier family 45 member 3 (SLC45A3), also known as prostate cancer-associated protein 6 or prostein, is a protein that in humans is encoded by the SLC45A3 gene.

Chromothripsis is a mutational process by which up to thousands of clustered chromosomal rearrangements occur in a single event in localised and confined genomic regions in one or a few chromosomes, and is known to be involved in both cancer and congenital diseases. It occurs through one massive genomic rearrangement during a single catastrophic event in the cell's history. It is believed that for the cell to be able to withstand such a destructive event, the occurrence of such an event must be the upper limit of what a cell can tolerate and survive. The chromothripsis phenomenon opposes the conventional theory that cancer is the gradual acquisition of genomic rearrangements and somatic mutations over time.
Genome instability refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or aneuploidy. Genome instability does occur in bacteria. In multicellular organisms genome instability is central to carcinogenesis, and in humans it is also a factor in some neurodegenerative diseases such as amyotrophic lateral sclerosis or the neuromuscular disease myotonic dystrophy.

Breakage-fusion-bridge (BFB) cycle is a mechanism of chromosomal instability, discovered by Barbara McClintock in the late 1930s.
Chromosomal instability (CIN) is a type of genomic instability in which chromosomes are unstable, such that either whole chromosomes or parts of chromosomes are duplicated or deleted. More specifically, CIN refers to the increase in rate of addition or loss of entire chromosomes or sections of them. The unequal distribution of DNA to daughter cells upon mitosis results in a failure to maintain euploidy leading to aneuploidy. In other words, the daughter cells do not have the same number of chromosomes as the cell they originated from. Chromosomal instability is the most common form of genetic instability and cause of aneuploidy.
Chimeric RNA, sometimes referred to as a fusion transcript, is composed of exons from two or more different genes that have the potential to encode novel proteins. These mRNAs are different from those produced by conventional splicing as they are produced by two or more gene loci.
The Cancer Genome Anatomy Project (CGAP), created by the National Cancer Institute (NCI) in 1997 and introduced by Al Gore, is an online database on normal, pre-cancerous and cancerous genomes. It also provides tools for viewing and analysis of the data, allowing for identification of genes involved in various aspects of tumor progression. The goal of CGAP is to characterize cancer at a molecular level by providing a platform with readily accessible updated data and a set of tools such that researchers can easily relate their findings to existing knowledge. There is also a focus on development of software tools that improve the usage of large and complex datasets. The project is directed by Daniela S. Gerhard, and includes sub-projects or initiatives, with notable ones including the Cancer Chromosome Aberration Project (CCAP) and the Genetic Annotation Initiative (GAI). CGAP contributes to many databases and organisations such as the NCBI contribute to CGAP's databases.
EWS/FLI1 is an oncogenic protein that is pathognomonic for Ewing sarcoma. It is found in approximately 90% of all Ewing sarcoma tumors with the remaining 10% of fusions substituting one fusion partner with a closely related family member.
Sclerosing epithelioid fibrosarcoma (SEF) is a very rare malignant tumor of soft tissues that on microscopic examination consists of small round or ovoid neoplastic epithelioid fibroblast-like cells, i.e. cells that have features resembling both epithelioid cells and fibroblasts. In 2020, the World Health Organization classified SEF as a distinct tumor type in the category of malignant fibroblastic and myofibroblastic tumors. However, current studies have reported that low-grade fibromyxoid sarcoma (LGFMS) has many clinically and pathologically important features characteristic of SEF; these studies suggest that LGSFMS may be an early form of, and over time progress to become, a SEF. Since the World Health Organization has classified LGFMS as one of the malignant fibroblastic and myofibroblastic tumors that is distinctly different than SEF, SEF and LGFMS are here regarded as different tumor forms.
The FET protein family consists of three similarly structured and functioning proteins. They and the genes in the FET gene family which encode them are: 1) the EWSR1 protein encoded by the EWSR1 gene located at band 12.2 of the long arm of chromosome 22; 2) the FUS protein encoded by the FUS gene located at band 16 on the short arm of chromosome 16; and 3) the TAF15 protein encoded by the TAF15 gene located at band 12 on the long arm of chromosome 7 The FET in this protein family's name derives from the first letters of FUS, EWSR1, and TAF15.
References
- 1 2 3 4 5 6 Baca, S. C., Prandi, D., Lawrence, M. S., Mosquera, J. M., Romanel, A., Drier, Y., ... Garraway, L. A. (2013). Punctuated evolution of prostate cancer genomes. Cell, 153(3), 666–77. doi:10.1016/j.cell.2013.03.021
- 1 2 Berger, M. F., Lawrence, M. S., Demichelis, F., Drier, Y., Cibulskis, K., Sivachenko, A. Y., ... Garraway, L. A. (2011). The genomic complexity of primary human prostate cancer. Nature, 470(7333), 214–20. doi:10.1038/nature09744
- 1 2 3 Shen, M. M. (2013). Chromoplexy: a new category of complex rearrangements in the cancer genome. Cancer Cell, 23(5), 567–9. doi:10.1016/j.ccr.2013.04.025
- ↑ Anderson ND, de Borja R, Young MD, Fuligni F, Rosic A, Roberts ND, Hajjar S, Layeghifard M, Novokmet A, Kowalski PE, Anaka M. Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science. 2018 Aug 31;361(6405):eaam8419.
- ↑ Pihan, G. a. (2013). Centrosome dysfunction contributes to chromosome instability, chromoanagenesis, and genome reprograming in cancer. Frontiers in Oncology, 3(November), 277. doi:10.3389/fonc.2013.00277
- ↑ Holland, A. J., & Cleveland, D. W. (2012). Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nature Medicine, 18(11), 1630–8. doi:10.1038/nm.2988
- ↑ Haffner, M. C., Aryee, M. J., Toubaji, A., Esopi, D. M., Albadine, R., Gurel, B., ... Yegnasubramanian, S. (2010). Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nature Genetics, 42(8), 668–75. doi:10.1038/ng.613
- ↑ Stephens, P. J., Greenman, C. D., Fu, B., Yang, F., Bignell, G. R., Mudie, L. J., ... Campbell, P. J. (2011). Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell, 144(1), 27–40. doi:10.1016/j.cell.2010.11.055
- 1 2 Forment, J. V., Kaidi, A., & Jackson, S. P. (2012). Chromothripsis and cancer: causes and consequences of chromosome shattering. Nature Reviews Cancer, 12 663-670.
- ↑ Zhang, C.-Z., Leibowitz, M. L., & Pellman, D. (2013). Chromothripsis and beyond: rapid genome evolution from complex chromosomal rearrangements. Genes & Development, 27(23), 2513–30. doi:10.1101/gad.229559.113