
Sourdough is a bread made by the fermentation of dough using wild lactobacillaceae and yeast. Lactic acid from fermentation imparts a sour taste and improves keeping qualities.

Lactobacillus is a genus of Gram-positive, aerotolerant anaerobes or microaerophilic, rod-shaped, non-spore-forming bacteria. Until 2020, the genus Lactobacillus comprised over 260 phylogenetically, ecologically, and metabolically diverse species; a taxonomic revision of the genus assigned lactobacilli to 25 genera.

Lactococcus is a genus of lactic acid bacteria that were formerly included in the genus Streptococcus Group N1. They are known as homofermenters meaning that they produce a single product, lactic acid in this case, as the major or only product of glucose fermentation. Their homofermentative character can be altered by adjusting environmental conditions such as pH, glucose concentration, and nutrient limitation. They are gram-positive, catalase-negative, non-motile cocci that are found singly, in pairs, or in chains. The genus contains strains known to grow at or below 7˚C.

Malolactic conversion is a process in winemaking in which tart-tasting malic acid, naturally present in grape must, is converted to softer-tasting lactic acid. Malolactic fermentation is most often performed as a secondary fermentation shortly after the end of the primary fermentation, but can sometimes run concurrently with it. The process is standard for most red wine production and common for some white grape varieties such as Chardonnay, where it can impart a "buttery" flavor from diacetyl, a byproduct of the reaction.
Lactiplantibacillus plantarum is a widespread member of the genus Lactiplantibacillus and commonly found in many fermented food products as well as anaerobic plant matter. L. plantarum was first isolated from saliva. Based on its ability to temporarily persist in plants, the insect intestine and in the intestinal tract of vertebrate animals, it was designated as a nomadic organism. L. plantarum is Gram positive, bacilli shaped bacterium. L. plantarum cells are rods with rounded ends, straight, generally 0.9–1.2 μm wide and 3–8 μm long, occurring singly, in pairs or in short chains. L. plantarum has one of the largest genomes known among the lactic acid bacteria and is a very flexible and versatile species. It is estimated to grow between pH 3.4 and 8.8. Lactiplantibacillus plantarum can grow in the temperature range 12 °C to 40 °C. The viable counts of the "L. plantarum" stored at refrigerated condition (4 °C) remained high, while a considerable reduction in the counts was observed stored at room temperature.
Pediococcus is a genus of gram-positive lactic acid bacteria, placed within the family of Lactobacillaceae. They usually occur in pairs or tetrads, and divide along two planes of symmetry, as do the other lactic acid cocci genera Aerococcus and Tetragenococcus. They are purely homofermentative. Pediococcus dextrinicus has recently been reassigned to the genus Lapidilactobacillus.

Lacticaseibacillus casei shirota is an organism that belongs to the largest genus in the family Lactobacillaceae, a lactic acid bacteria (LAB), that was previously classified as Lactobacillus casei-01. This bacteria has been identified as facultatively anaerobic or microaerophilic, acid-tolerant, non-spore-forming bacteria. The taxonomy of this group has been debated for several years because researchers struggled to differentiate between the strains of L. casei and L. paracasei. It has recently been accepted as a single species with five subspecies: L. casei subsp. rhamnosus, L. casei subsp. alactosus, L. casei subsp. casei, L. casei subsp. tolerans, and L. casei subsp. pseudoplantarum. The taxonomy of this genus was determined according to the phenotypic, physiological, and biochemical similarities.
Leuconostoc is a genus of gram-positive bacteria, placed within the family of Lactobacillaceae. They are generally avoid cocci often forming chains. Leuconostoc spp. are intrinsically resistant to vancomycin and are catalase-negative. All species within this genus are heterofermentative and are able to produce dextran from sucrose. They are generally slime-forming.

Lactobacillales are an order of gram-positive, low-GC, acid-tolerant, generally nonsporulating, nonrespiring, either rod-shaped (bacilli) or spherical (cocci) bacteria that share common metabolic and physiological characteristics. These bacteria, usually found in decomposing plants and milk products, produce lactic acid as the major metabolic end product of carbohydrate fermentation, giving them the common name lactic acid bacteria (LAB).

The Lactobacillaceae are a family of lactic acid bacteria. It is the only family in the lactic acid bacteria which includes homofermentative and heterofermentative organisms; in the Lactobacillaceae, the pathway used for hexose fermentation is a genus-specific trait. Lactobacillaceae include the homofermentative lactobacilli Lactobacillus, Holzapfelia, Amylolactobacillus, Bombilactobacillus, Companilactobacillus, Lapidilactobacillus, Agrilactobacillus, Schleiferilactobacillus, Loigolactobacillus, Lacticaseibacillus, Latilactobacillus, Dellaglioa, Liquorilactobacillus, Ligilactobacillus, and Lactiplantibacillus; the heterofermentative lactobacilli Furfurilactobacillus, Paucilactobacillus, Limosilactobacillus, Fructilactobacillus, Acetilactobacillus, Apilactobacillus, Levilactobacillus, Secundilactobacillus, and Lentilactobacillus, which were previously classified in the genus Lactobacillus; and the heterofermentative genera Convivina, Fructobacillus, Leuconostoc, Oenococcus, and Weissella which were previously classified in the Leuconostocaceae.

Dadiah (Minangkabau) or dadih (Indonesian) a traditional fermented milk popular among people of West Sumatra, Indonesia, is made by pouring fresh, raw, unheated, buffalo milk into a bamboo tube capped with a banana leaf and allowing it to ferment spontaneously at room temperature for two days.
Leuconostoc mesenteroides is a species of lactic acid bacteria associated with fermentation, under conditions of salinity and low temperatures. In some cases of vegetable and food storage, it was associated with pathogenicity. L. mesenteroides is approximately 0.5-0.7 µm in diameter and has a length of 0.7-1.2 µm, producing small grayish colonies that are typically less than 1.0 mm in diameter. It is facultatively anaerobic, Gram-positive, non-motile, non-sporogenous, and spherical. It often forms lenticular coccoid cells in pairs and chains, however, it can occasionally forms short rods with rounded ends in long chains, as its shape can differ depending on what media the species is grown on. L. mesenteroides grows best at 30°C, but can survive in temperatures ranging from 10°C to 30°C. Its optimum pH is 5.5, but can still show growth in pH of 4.5-7.0.
Lentilactobacillus buchneri is a gram-positive, non-spore forming, anaerobic, rod prokaryote. L. buchneri is a heterofermentative bacteria that produces lactic acid and acetic acid during fermentation. It is used as a bacterial inoculant to improve the aerobic stability of silage. These bacteria are inoculated and used for preventing heating and spoilage after exposure to air.
Sakacins are bacteriocins produced by Lactobacillus sakei. They are often clustered with the other lactic acid bacteriocins. The best known sakacins are sakacin A, G, K, P, and Q. In particular, sakacin A and P have been well characterized.

Latilactobacillus sakei is the type species of the genus Latilactobacillus that was previously classified in the genus Lactobacillus. It is homofermentative; hexoses are metabolized via Glycolysis to lactic acid as main metabolite; pentoses are fermented via the Phosphoketolase pathway to lactic and acetic acids.
Lactiplantibacillus paraplantarum is a rod-shaped species of lactic acid bacteria first isolated from beer and human faeces. It is facultatively heterofermentative. Strain CNRZ 1885 is the type strain.
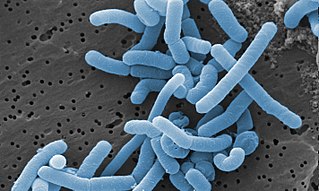
Lacticaseibacillus paracasei is a gram-positive, homofermentative species of lactic acid bacteria that are commonly used in dairy product fermentation and as probiotic cultures. Lc. paracasei is a bacterium that operates by commensalism. It is commonly found in many human habitats such as human intestinal tracts and mouths as well as sewages, silages, and previously mentioned dairy products. The name includes morphology, a rod-shaped bacterium with a width of 2.0 to 4.0μm and length of 0.8 to 1.0μm.
Limosilactobacillus pontis is a rod-shaped, Gram-positive facultatively anaerobic bacterium. Along with other Lactobacillus species, it is capable of converting sugars, such as lactose, into lactic acid. Limosilactobacillus pontis is classified under the phylum Bacillota, class Bacilli, and is a member of the family Lactobacillaceae and is found to be responsible for the fermentation of sourdough, along with many other Lactobacillus species. This microorganism produces lactic acid during the process of fermentation, which gives sourdough bread its characteristic sour taste.
Fructilactobacillus fructivorans is a gram-positive bacteria and a member of the genus Fructilactobacillus in the family Lactobacillaceae. It is found in wine, beer, grape must, dairy, sauerkraut, meat, and fish. They are facultative anaerobics and experience best growth in environments with 5-10% CO2. Temperature for growth is between 2 °C and 53 °C, with the optimum temperature between 30 °C and 40 °C and a pH level between 5.5 and 6.2. The bacterium is rod shaped and can be found in the following forms: single, pairs, chains of varying lengths, or long curved filaments. Lactobacillus fructivorans is non-motile. The main end product of the metabolic process is lactate, although ethanol, acetate, formate, CO2, and succinate may also be produced.
Lactiplantibacillus is a genus of lactic acid bacteria.








