
Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two hybridized nucleic acid strands, using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.

DNA polymerase I is an enzyme that participates in the process of prokaryotic DNA replication. Discovered by Arthur Kornberg in 1956, it was the first known DNA polymerase. It was initially characterized in E. coli and is ubiquitous in prokaryotes. In E. coli and many other bacteria, the gene that encodes Pol I is known as polA. The E. coli Pol I enzyme is composed of 928 amino acids, and is an example of a processive enzyme — it can sequentially catalyze multiple polymerisation steps without releasing the single-stranded template. The physiological function of Pol I is mainly to support repair of damaged DNA, but it also contributes to connecting Okazaki fragments by deleting RNA primers and replacing the ribonucleotides with DNA.
dnaQ is the gene encoding the ε subunit of DNA polymerase III in Escherichia coli. The ε subunit is one of three core proteins in the DNA polymerase complex. It functions as a 3’→5’ DNA directed proofreading exonuclease that removes incorrectly incorporated bases during replication. dnaQ may also be referred to as mutD.
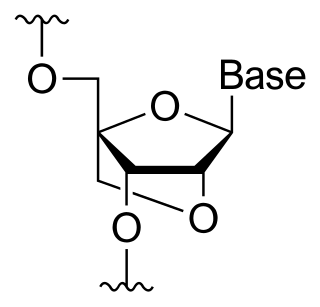
A locked nucleic acid (LNA), also known as bridged nucleic acid (BNA), and often referred to as inaccessible RNA, is a modified RNA nucleotide in which the ribose moiety is modified with an extra bridge connecting the 2' oxygen and 4' carbon. The bridge "locks" the ribose in the 3'-endo (North) conformation, which is often found in the A-form duplexes. This structure provides for increased stability against enzymatic degradation. LNA also offers improved specificity and affinity in base-pairing as a monomer or a constituent of an oligonucleotide. LNA nucleotides can be mixed with DNA or RNA residues in a oligonucleotide.

DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for single- or double-stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that identify a base pair. However, there are some known minor groove DNA-binding ligands such as netropsin, distamycin, Hoechst 33258, pentamidine, DAPI and others.

Werner syndrome ATP-dependent helicase, also known as DNA helicase, RecQ-like type 3, is an enzyme that in humans is encoded by the WRN gene. WRN is a member of the RecQ Helicase family. Helicase enzymes generally unwind and separate double-stranded DNA. These activities are necessary before DNA can be copied in preparation for cell division. Helicase enzymes are also critical for making a blueprint of a gene for protein production, a process called transcription. Further evidence suggests that Werner protein plays a critical role in repairing DNA. Overall, this protein helps maintain the structure and integrity of a person's DNA.
Flap endonucleases are a class of nucleolytic enzymes that act as both 5'-3' exonucleases and structure-specific endonucleases on specialised DNA structures that occur during the biological processes of DNA replication, DNA repair, and DNA recombination. Flap endonucleases have been identified in eukaryotes, prokaryotes, archaea, and some viruses. Organisms can have more than one FEN homologue; this redundancy may give an indication of the importance of these enzymes. In prokaryotes, the FEN enzyme is found as an N-terminal domain of DNA polymerase I, but some prokaryotes appear to encode a second homologue.

DNA-(apurinic or apyrimidinic site) lyase is an enzyme that in humans is encoded by the APEX1 gene.

Replication protein A 70 kDa DNA-binding subunit is a protein that in humans is encoded by the RPA1 gene.

DNA polymerase subunit gamma is an enzyme that in humans is encoded by the POLG gene. Mitochondrial DNA polymerase is heterotrimeric, consisting of a homodimer of accessory subunits plus a catalytic subunit. The protein encoded by this gene is the catalytic subunit of mitochondrial DNA polymerase. Defects in this gene are a cause of progressive external ophthalmoplegia with mitochondrial DNA deletions 1 (PEOA1), sensory ataxic neuropathy dysarthria and ophthalmoparesis (SANDO), Alpers-Huttenlocher syndrome (AHS), and mitochondrial neurogastrointestinal encephalopathy syndrome (MNGIE).

Origin recognition complex subunit 2 is a protein that is encoded by the ORC2 (ORC2L) gene in humans.

Twinkle protein also known as twinkle mtDNA helicase is a mitochondrial protein that in humans is encoded by the TWNK gene located in the long arm of chromosome 10 (10q24.31).

Three prime repair exonuclease 2 is an enzyme that in humans is encoded by the TREX2 gene.

DNA replication licensing factor MCM8 is a protein that in humans is encoded by the MCM8 gene.
Φ29 DNA polymerase is an enzyme from the bacteriophage Φ29. It is being increasingly used in molecular biology for multiple displacement DNA amplification procedures, and has a number of features that make it particularly suitable for this application. It was discovered and characterised by Spanish scientists Luis Blanco and Margarita Salas.

PrimPol is a protein encoded by the PRIMPOL gene in humans. PrimPol is a eukaryotic protein with both DNA polymerase and DNA Primase activities involved in translesion DNA synthesis. It is the first eukaryotic protein to be identified with priming activity using deoxyribonucleotides. It is also the first protein identified in the mitochondria to have translesion DNA synthesis activities.

DNA polymerase alpha subunit 2 is an enzyme that in humans is encoded by the POLA2 gene.

DNA polymerase alpha catalytic subunit is an enzyme that in humans is encoded by the POLA1 gene.

Replication protein A 32 kDa subunit is a protein that in humans is encoded by the RPA2 gene.




















