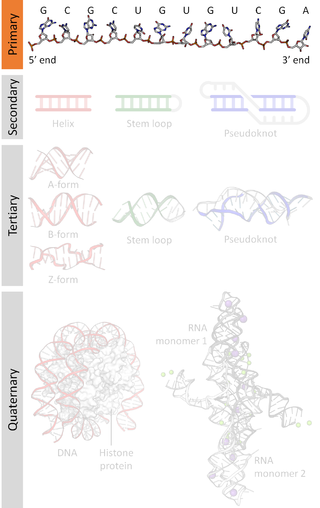
A nucleic acid sequence is a succession of bases within the nucleotides forming alleles within a DNA or RNA (GACU) molecule. This succession is denoted by a series of a set of five different letters that indicate the order of the nucleotides. By convention, sequences are usually presented from the 5' end to the 3' end. For DNA, with its double helix, there are two possible directions for the notated sequence; of these two, the sense strand is used. Because nucleic acids are normally linear (unbranched) polymers, specifying the sequence is equivalent to defining the covalent structure of the entire molecule. For this reason, the nucleic acid sequence is also termed the primary structure.
The 5′ untranslated region is the region of a messenger RNA (mRNA) that is directly upstream from the initiation codon. This region is important for the regulation of translation of a transcript by differing mechanisms in viruses, prokaryotes and eukaryotes. While called untranslated, the 5′ UTR or a portion of it is sometimes translated into a protein product. This product can then regulate the translation of the main coding sequence of the mRNA. In many organisms, however, the 5′ UTR is completely untranslated, instead forming a complex secondary structure to regulate translation.
The Shine–Dalgarno (SD) sequence is a ribosomal binding site in bacterial and archaeal messenger RNA, generally located around 8 bases upstream of the start codon AUG. The RNA sequence helps recruit the ribosome to the messenger RNA (mRNA) to initiate protein synthesis by aligning the ribosome with the start codon. Once recruited, tRNA may add amino acids in sequence as dictated by the codons, moving downstream from the translational start site.
In molecular biology, small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, the C/D box snoRNAs, which are associated with methylation, and the H/ACA box snoRNAs, which are associated with pseudouridylation. SnoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNAs that direct RNA editing in trypanosomes or the guide RNAs (gRNAs) used by Cas9 for CRISPR gene editing.

In molecular genetics, an untranslated region refers to either of two sections, one on each side of a coding sequence on a strand of mRNA. If it is found on the 5' side, it is called the 5' UTR, or if it is found on the 3' side, it is called the 3' UTR. mRNA is RNA that carries information from DNA to the ribosome, the site of protein synthesis (translation) within a cell. The mRNA is initially transcribed from the corresponding DNA sequence and then translated into protein. However, several regions of the mRNA are usually not translated into protein, including the 5' and 3' UTRs.

The msiK RNA motif describes a conserved RNA structure discovered using bioinformatics. The RNA is always found in the presumed 5' untranslated regions of genes annotated as msiK, and is therefore hypothesized to be an RNA-based cis-regulatory element that regulates these genes.

An RNA thermometer is a temperature-sensitive non-coding RNA molecule which regulates gene expression. Its unique characteristic it is that it does not need proteins or metabolites to function, but only reacts to temperature changes. RNA thermometers often regulate genes required during either a heat shock or cold shock response, but have been implicated in other regulatory roles such as in pathogenicity and starvation.

The aspS RNA motif is a conserved RNA structure that was discovered by bioinformatics. aspS motifs are found in a specific lineage of Actinomycetota.
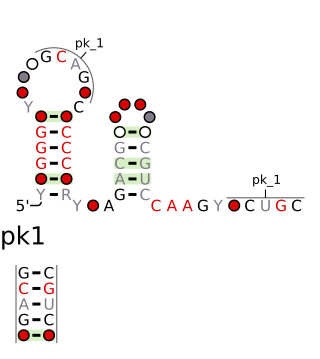
The chrB-a RNA motif and chrB-b RNA motif refer to a related, conserved RNA structure that was discovered by bioinformatics. The structures of these motifs are similar, and some genomic locations are predicted to exhibit both motifs. The chrB-b motif has an extra pseudoknot that is not consistently found in chrB-a examples. It was proposed that the two motifs could be unified into one common structure, with additional information.

The DUF2800 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF2800 motif RNAs are found in Bacillota. DUF2800 RNAs are also predicted in the phyla Actinomycetota and Synergistota, although these RNAs are likely the result of recent horizontal gene transfer or conceivably sequence contamination.

The DUF3577 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF3577 motifs are found in the organism Cardiobacterium valvarum and metagenomic sequences from unknown organisms.
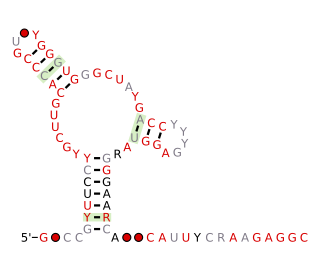
The Fibro-purF RNA motif is a conserved RNA structure that was discovered by bioinformatics. Fibro-purF motif RNAs are found in Fibrobacterota, a group of bacteria that are common in cow rumen. Additionally, the RNAs are found in metagenomic sequences of DNA isolated from cow rumen.

The folE RNA motif, now known as the THF-II riboswitch, is a conserved RNA structure that was discovered by bioinformatics. folE motifs are found in Alphaproteobacteria.
IS605-orfB RNA motifs refer to conserved RNA or DNA structures that were discovered by bioinformatics. Although such motifs were published as a RNA candidates, there is some reason to suspect that they might function as a single-stranded DNA. In terms of secondary structure, RNA and DNA are difficult to distinguish when only sequence information is available. If the motifs function as RNA, they likely are small RNAs, that are independently transcribed.
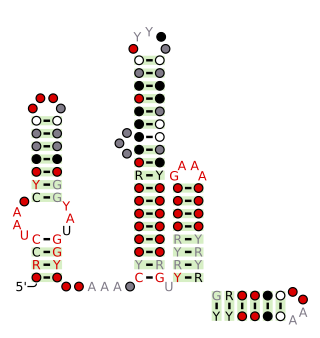
The Latescibacteria, OD1, OP11, TM7 RNA motif is a conserved RNA structure that was discovered by bioinformatics. LOOT motif RNAs are found in multiple bacterial phyla that have only recently been discovered, and are currently not well understood: Latescibacteria, OD1/Parcubacteria, OP11 AND TM7. In some cases, no specific organism has been isolated in the relevant phylum, but the existence of the bacterial phylum is known only through analysis of metagenomic sequences. Curiously, the LOOT motif is not known in any phylum that has been studied for a long time.
lysM RNA motifs are conserved RNA structures that were discovered by bioinformatics. Such bacterial motifs are defined by consistently being upstream of 'lysM' genes, which encode lysin protein domains, a conserved domain that participates in cell wall degradation. lysM motif RNAs likely function as cis-regulatory elements, in view of their positions upstream of protein-coding genes, although this hypothesis is not certain.

The osmY RNA motif is a conserved RNA structure that was discovered by bioinformatics. osmY motif RNAs are found in Enterobacteriaceae organisms, although it is not predicted to reside in Escherichia coli.

The PGK RNA motif is a conserved RNA structure that was discovered by bioinformatics. PGK motif RNAs are found in metagenomic sequences isolated from the gastrointestinal tract of mammals. PGK RNAs have not yet been detected in a classified organism.
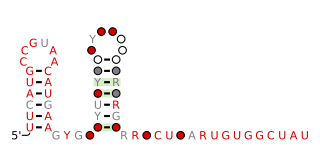
The queA RNA motif is a conserved RNA structure that was discovered by bioinformatics. queA motif RNAs have not yet been found in any classified organism; they are known from metagenomic sequences.
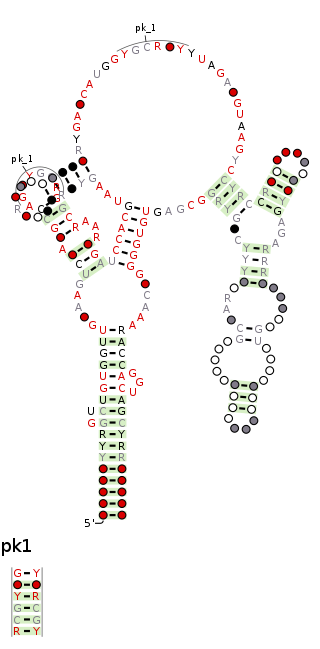
The raiA RNA motif is a conserved RNA structure that was discovered by bioinformatics. raiA motif RNAs are found in Actinomycetota and Bacillota, and have many conserved features—including conserved nucleotide positions, conserved secondary structures and associated protein-coding genes—in both of these phyla. Some conserved features of the raiA RNA motif suggest that they function as cis-regulatory elements, but other aspects of the motif suggest otherwise.

















