
Bioinformatics is an interdisciplinary field of science that develops methods and software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, chemistry, physics, computer science, computer programming, information engineering, mathematics and statistics to analyze and interpret biological data. The subsequent process of analyzing and interpreting data is referred to as computational biology.

Neurofeedback is a form of biofeedback that uses electrical potentials in the brain to reinforce desired brain states through operant conditioning. This process is non-invasive and typically collects brain activity data using electroencephalography (EEG). Several neurofeedback protocols exist, with potential additional benefit from use of quantitative electroencephalography (QEEG) or functional magnetic resonance imaging (fMRI) to localize and personalize treatment. Related technologies include functional near-infrared spectroscopy-mediated (fNIRS) neurofeedback, hemoencephalography biofeedback (HEG), and fMRI biofeedback.
BioJava is an open-source software project dedicated to provide Java tools to process biological data. BioJava is a set of library functions written in the programming language Java for manipulating sequences, protein structures, file parsers, Common Object Request Broker Architecture (CORBA) interoperability, Distributed Annotation System (DAS), access to AceDB, dynamic programming, and simple statistical routines. BioJava supports a huge range of data, starting from DNA and protein sequences to the level of 3D protein structures. The BioJava libraries are useful for automating many daily and mundane bioinformatics tasks such as to parsing a Protein Data Bank (PDB) file, interacting with Jmol and many more. This application programming interface (API) provides various file parsers, data models and algorithms to facilitate working with the standard data formats and enables rapid application development and analysis.

The superior temporal gyrus (STG) is one of three gyri in the temporal lobe of the human brain, which is located laterally to the head, situated somewhat above the external ear.
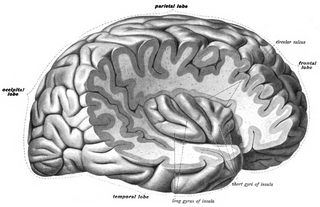
The insular cortex is a portion of the cerebral cortex folded deep within the lateral sulcus within each hemisphere of the mammalian brain.
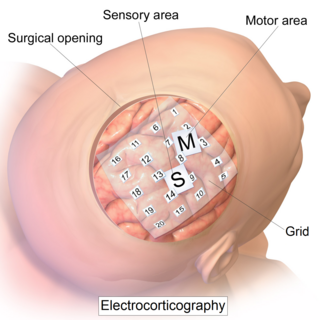
Electrocorticography (ECoG), a type of intracranial electroencephalography (iEEG), is a type of electrophysiological monitoring that uses electrodes placed directly on the exposed surface of the brain to record electrical activity from the cerebral cortex. In contrast, conventional electroencephalography (EEG) electrodes monitor this activity from outside the skull. ECoG may be performed either in the operating room during surgery or outside of surgery. Because a craniotomy is required to implant the electrode grid, ECoG is an invasive procedure.

ImageJ is a Java-based image processing program developed at the National Institutes of Health and the Laboratory for Optical and Computational Instrumentation. Its first version, ImageJ 1.x, is developed in the public domain, while ImageJ2 and the related projects SciJava, ImgLib2, and SCIFIO are licensed with a permissive BSD-2 license. ImageJ was designed with an open architecture that provides extensibility via Java plugins and recordable macros. Custom acquisition, analysis and processing plugins can be developed using ImageJ's built-in editor and a Java compiler. User-written plugins make it possible to solve many image processing and analysis problems, from three-dimensional live-cell imaging to radiological image processing, multiple imaging system data comparisons to automated hematology systems. ImageJ's plugin architecture and built-in development environment has made it a popular platform for teaching image processing.
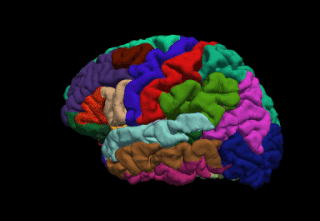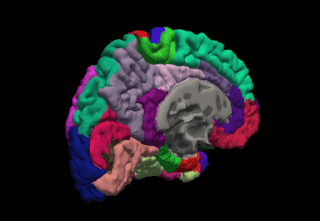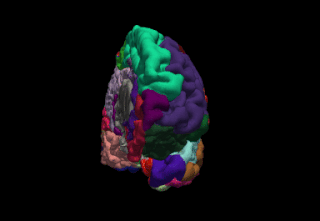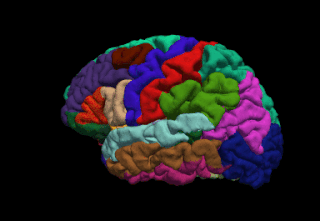
FreeSurfer is a brain imaging software package originally developed by Bruce Fischl, Anders Dale, Martin Sereno, and Doug Greve. Development and maintenance of FreeSurfer is now the primary responsibility of the Laboratory for Computational Neuroimaging at the Athinoula A. Martinos Center for Biomedical Imaging. FreeSurfer contains a set of programs with a common focus of analyzing magnetic resonance imaging (MRI) scans of brain tissue. It is an important tool in functional brain mapping and contains tools to conduct both volume based and surface based analysis. FreeSurfer includes tools for the reconstruction of topologically correct and geometrically accurate models of both the gray/white and pial surfaces, for measuring cortical thickness, surface area and folding, and for computing inter-subject registration based on the pattern of cortical folds.
Epilepsy surgery involves a neurosurgical procedure where an area of the brain involved in seizures is either resected, ablated, disconnected or stimulated. The goal is to eliminate seizures or significantly reduce seizure burden. Approximately 60% of all people with epilepsy have focal epilepsy syndromes. In 15% to 20% of these patients, the condition is not adequately controlled with anticonvulsive drugs. Such patients are potential candidates for surgical epilepsy treatment.
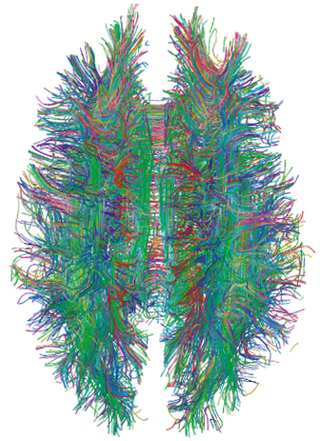
A connectome is a comprehensive map of neural connections in the brain, and may be thought of as its "wiring diagram". An organism's nervous system is made up of neurons which communicate through synapses. A connectome is constructed by tracing the neuron in a nervous system and mapping where neurons are connected through synapses.
Connectomics is the production and study of connectomes: comprehensive maps of connections within an organism's nervous system. More generally, it can be thought of as the study of neuronal wiring diagrams with a focus on how structural connectivity, individual synapses, cellular morphology, and cellular ultrastructure contribute to the make up of a network. The nervous system is a network made of billions of connections and these connections are responsible for our thoughts, emotions, actions, memories, function and dysfunction. Therefore, the study of connectomics aims to advance our understanding of mental health and cognition by understanding how cells in the nervous system are connected and communicate. Because these structures are extremely complex, methods within this field use a high-throughput application of functional and structural neural imaging, most commonly magnetic resonance imaging (MRI), electron microscopy, and histological techniques in order to increase the speed, efficiency, and resolution of these nervous system maps. To date, tens of large scale datasets have been collected spanning the nervous system including the various areas of cortex, cerebellum, the retina, the peripheral nervous system and neuromuscular junctions.
Brain morphometry is a subfield of both morphometry and the brain sciences, concerned with the measurement of brain structures and changes thereof during development, aging, learning, disease and evolution. Since autopsy-like dissection is generally impossible on living brains, brain morphometry starts with noninvasive neuroimaging data, typically obtained from magnetic resonance imaging (MRI). These data are born digital, which allows researchers to analyze the brain images further by using advanced mathematical and statistical methods such as shape quantification or multivariate analysis. This allows researchers to quantify anatomical features of the brain in terms of shape, mass, volume, and to derive more specific information, such as the encephalization quotient, grey matter density and white matter connectivity, gyrification, cortical thickness, or the amount of cerebrospinal fluid. These variables can then be mapped within the brain volume or on the brain surface, providing a convenient way to assess their pattern and extent over time, across individuals or even between different biological species. The field is rapidly evolving along with neuroimaging techniques—which deliver the underlying data—but also develops in part independently from them, as part of the emerging field of neuroinformatics, which is concerned with developing and adapting algorithms to analyze those data.

Epilepsy is a neurological condition of recurrent episodes of unprovoked epileptic seizures. A seizure is an abnormal neuronal brain activity that can cause intellectual, emotional, and social consequences. Epilepsy affects children and adults of all ages and races, it is one of the most common neurological disorders of the nervous system. As well as, this condition is more common among children than adults affecting about 6 out of 1000 US children that are between the age of 0 to 5 years old. The epileptic seizures can be of different types depending on the part of the brain that was affected, seizures are classified in 2 main types partial seizure or genralized seizure.

Resting state fMRI is a method of functional magnetic resonance imaging (fMRI) that is used in brain mapping to evaluate regional interactions that occur in a resting or task-negative state, when an explicit task is not being performed. A number of resting-state brain networks have been identified, one of which is the default mode network. These brain networks are observed through changes in blood flow in the brain which creates what is referred to as a blood-oxygen-level dependent (BOLD) signal that can be measured using fMRI.

CONN is a Matlab-based cross-platform imaging software for the computation, display, and analysis of functional connectivity in fMRI in the resting state and during task.
Dynamic causal modeling (DCM) is a framework for specifying models, fitting them to data and comparing their evidence using Bayesian model comparison. It uses nonlinear state-space models in continuous time, specified using stochastic or ordinary differential equations. DCM was initially developed for testing hypotheses about neural dynamics. In this setting, differential equations describe the interaction of neural populations, which directly or indirectly give rise to functional neuroimaging data e.g., functional magnetic resonance imaging (fMRI), magnetoencephalography (MEG) or electroencephalography (EEG). Parameters in these models quantify the directed influences or effective connectivity among neuronal populations, which are estimated from the data using Bayesian statistical methods.
Catherine J. "Cathy" Price is a British neuroscientist and academic. She is a professor of cognitive neuroscience and director of the Wellcome Trust Centre for Neuroimaging at University College London.












