Related Research Articles
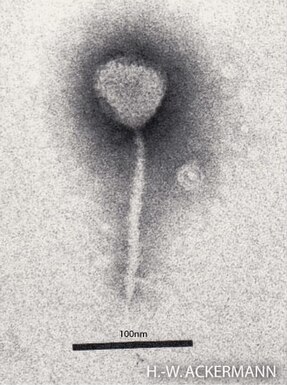
Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

FtsZ is a protein encoded by the ftsZ gene that assembles into a ring at the future site of bacterial cell division. FtsZ is a prokaryotic homologue of the eukaryotic protein tubulin. The initials FtsZ mean "Filamenting temperature-sensitive mutant Z." The hypothesis was that cell division mutants of E. coli would grow as filaments due to the inability of the daughter cells to separate from one another. FtsZ is found in almost all bacteria, many archaea, all chloroplasts and some mitochondria, where it is essential for cell division. FtsZ assembles the cytoskeletal scaffold of the Z ring that, along with additional proteins, constricts to divide the cell in two.
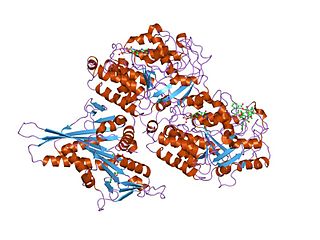
Tubulin in molecular biology can refer either to the tubulin protein superfamily of globular proteins, or one of the member proteins of that superfamily. α- and β-tubulins polymerize into microtubules, a major component of the eukaryotic cytoskeleton. Microtubules function in many essential cellular processes, including mitosis. Tubulin-binding drugs kill cancerous cells by inhibiting microtubule dynamics, which are required for DNA segregation and therefore cell division.

The SOS response is a global response to DNA damage in which the cell cycle is arrested and DNA repair and mutagenesis is induced. The system involves the RecA protein. The RecA protein, stimulated by single-stranded DNA, is involved in the inactivation of the repressor (LexA) of SOS response genes thereby inducing the response. It is an error-prone repair system that contributes significantly to DNA changes observed in a wide range of species.

The nucleoid is an irregularly shaped region within the prokaryotic cell that contains all or most of the genetic material. The chromosome of a prokaryote is circular, and its length is very large compared to the cell dimensions, so it needs to be compacted in order to fit. In contrast to the nucleus of a eukaryotic cell, it is not surrounded by a nuclear membrane. Instead, the nucleoid forms by condensation and functional arrangement with the help of chromosomal architectural proteins and RNA molecules as well as DNA supercoiling. The length of a genome widely varies and a cell may contain multiple copies of it.
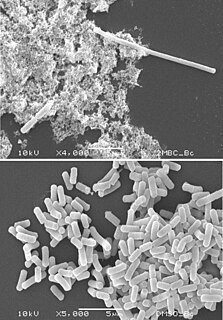
Filamentation is the anomalous growth of certain bacteria, such as Escherichia coli, in which cells continue to elongate but do not divide. The cells that result from elongation without division have multiple chromosomal copies. In the absence of antibiotics or other stressors, filamentation occurs at a low frequency in bacterial populations, the increased cell length protecting bacteria from protozoan predation and neutrophil phagocytosis by making ingestion of the cells more difficult. Filamentation is also thought to protect bacteria from antibiotics, and is associated with other aspects of bacterial virulence such as biofilm formation. The number and length of filaments within a bacterial population increases when the bacteria are treated with various chemical and physical agents. Some of the key genes involved in filamentation in E. coli include sulA and minCD.
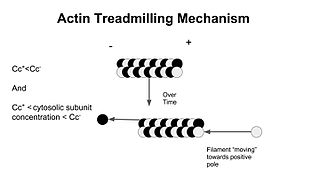
Treadmilling is a phenomenon observed in many cellular cytoskeletal filaments, especially in actin filaments and microtubules. It occurs when one end of a filament grows in length while the other end shrinks resulting in a section of filament seemingly "moving" across a stratum or the cytosol. This is due to the constant removal of the protein subunits from these filaments at one end of the filament while protein subunits are constantly added at the other end. Treadmilling was discovered by Wegner, who defined the thermodynamic and kinetic constraints. Wegner recognized that: “The equilibrium constant (K) for association of a monomer with a polymer is the same at both ends, since the addition of a monomer to each end leads to the same polymer.”; a simple reversible polymer can’t treadmill; ATP hydrolysis is required. GTP is hydrolyzed for microtubule treadmilling.
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.
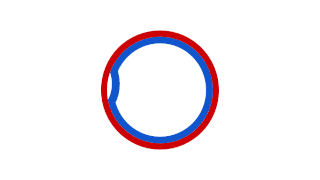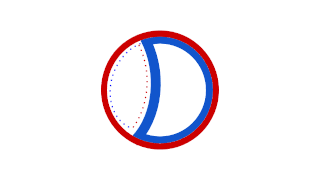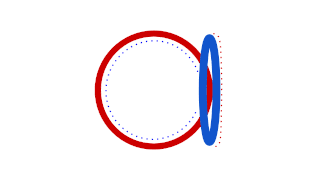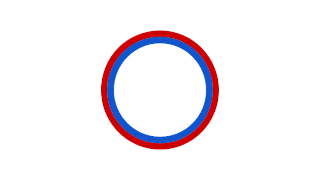
Prokaryotic DNA Replication is the process by which a prokaryote duplicates its DNA into another copy that is passed on to daughter cells. Although it is often studied in the model organism E. coli, other bacteria show many similarities. Replication is bi-directional and originates at a single origin of replication (OriC). It consists of three steps: Initiation, elongation, and termination.

DicF RNA is a non-coding RNA that is an antisense inhibitor of cell division gene ftsZ. DicF is bound by the Hfq protein which enhances its interaction with its targets. Pathogenic E. coli strains possess multiple copies of sRNA DicF in their genomes, while non-pathogenic strains do not. DicF and Hfq are both necessary to reduce FtsZ protein levels, leading to cell filamentation under anaerobic conditions.
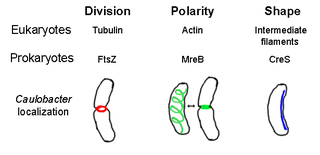
The prokaryotic cytoskeleton is the collective name for all structural filaments in prokaryotes. It was once thought that prokaryotic cells did not possess cytoskeletons, but advances in visualization technology and structure determination led to the discovery of filaments in these cells in the early 1990s. Not only have analogues for all major cytoskeletal proteins in eukaryotes been found in prokaryotes, cytoskeletal proteins with no known eukaryotic homologues have also been discovered. Cytoskeletal elements play essential roles in cell division, protection, shape determination, and polarity determination in various prokaryotes.
Fission, in biology, is the division of a single entity into two or more parts and the regeneration of those parts to separate entities resembling the original. The object experiencing fission is usually a cell, but the term may also refer to how organisms, bodies, populations, or species split into discrete parts. The fission may be binary fission, in which a single organism produces two parts, or multiple fission, in which a single entity produces multiple parts.
Bacterial morphological plasticity refers to changes in the shape and size that bacterial cells undergo when they encounter stressful environments. Although bacteria have evolved complex molecular strategies to maintain their shape, many are able to alter their shape as a survival strategy in response to protist predators, antibiotics, the immune response, and other threats.

The Min System is a mechanism composed of three proteins MinC, MinD, and MinE used by E. coli as a means of properly localizing the septum prior to cell division. Each component participates in generating a dynamic oscillation of FtsZ protein inhibition between the two bacterial poles to precisely specify the mid-zone of the cell, allowing the cell to accurately divide in two. This system is known to function in conjunction with a second negative regulatory system, the nucleoid occlusion system (NO), to ensure proper spatial and temporal regulation of chromosomal segregation and division.
The MinD protein is one of three proteins encoded by the minB operon and also a part of the ParA family of ATPases. It is required to generate pole to pole oscillations prior to bacterial cell division as a means of specifying the midzone of the cell. It is a peripheral membrane ATPase involved in plasmid partitioning.
The MinE protein is one of three proteins of the Min system encoded by the minB operon required to generate pole to pole oscillations prior to bacterial cell division as a means of specifying the midzone of the cell, as seen in E.coli.

FtsA is a bacterial protein that is related to actin by overall structural similarity and in its ATP binding pocket.
The bacterial murein precursor exporter (MPE) family is a member of the cation diffusion facilitator (CDF) superfamily of membrane transporters. Members of the MPE family are found in a large variety of Gram-negative and Gram-positive bacteria and facilitate the translocation of lipid-linked murein precursors. A representative list of proteins belonging to the MPE family can be found in the Transporter Classification Database.

The divisome is a protein complex in bacteria that is responsible for cell division, constriction of inner and outer membranes during division, and peptidoglycan (PG) synthesis at the division site. The divisome is a membrane protein complex with proteins on both sides of the cytoplasmic membrane. In gram-negative cells it is located in the inner membrane. The divisome is nearly ubiquitous in bacteria although its composition may vary between species.
In bacteriology, minicells are bacterial cells that are smaller than usual. The first minicells reported were from a strain of Escherichia coli that had a mutation in the Min System that lead to mis-localization of the septum during cell division and the production of cells of random sizes.
References
- ↑ Pichoff S, Lutkenhaus J (2001). "Escherichia coli division inhibitor MinCD blocks separation by preventing Z-ring formation". Journal of Bacteriology. 183 (22): 6630–6635. doi:10.1128/JB.183.22.6630-6635.2001. PMC 95494 . PMID 11673433.
- ↑ de Boer PA, Crossley RE, Rothfield LI (1989). "A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli". Cell. 56 (4): 641–9. doi:10.1016/0092-8674(89)90586-2. PMID 2645057. S2CID 7650379.
- ↑ Raskin DM, de Boer PA (1999). "Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli". Proceedings of the National Academy of Sciences of the United States of America. 96 (9): 4971–6. doi: 10.1073/pnas.96.9.4971 . PMC 21801 . PMID 10220403.
- ↑ Rowlett VW, Margolin W (2015). "The Min system and other nucleoid-independent regulators of Z ring positioning". Frontiers in Microbiology. 6: 478. doi: 10.3389/fmicb.2015.00478 . PMC 4429545 . PMID 26029202.
- ↑ Suzanne C. Cordell; Rebecca E. Anderson & Jan Löwe (2001). "Crystal structure of the bacterial cell division inhibitor MinC". The EMBO Journal. 20 (10): 2454–2461. doi:10.1093/emboj/20.10.2454. PMC 125452 . PMID 11350934.
- ↑ Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J (1999). "The MinC Component of the Division Site Selection System in Escherichia Coli interacts with FtsZ to Prevent Polymerization". PNAS. 96 (26): 14819–14824. doi: 10.1073/pnas.96.26.14819 . PMC 24731 . PMID 10611296.