
The cytoskeleton is a complex, dynamic network of interlinking protein filaments present in the cytoplasm of all cells, including bacteria and archaea. It extends from the cell nucleus to the cell membrane and is composed of similar proteins in the various organisms. In eukaryotes, it is composed of three main components, microfilaments, intermediate filaments and microtubules, and these are all capable of rapid growth or disassembly dependent on the cell's requirements.

Intermediate filaments (IFs) are cytoskeletal structural components found in the cells of vertebrates, and many invertebrates. Homologues of the IF protein have been noted in an invertebrate, the cephalochordate Branchiostoma.

FtsZ is a protein encoded by the ftsZ gene that assembles into a ring at the future site of bacterial cell division. FtsZ is a prokaryotic homologue of the eukaryotic protein tubulin. The initials FtsZ mean "Filamenting temperature-sensitive mutant Z." The hypothesis was that cell division mutants of E. coli would grow as filaments due to the inability of the daughter cells to separate from one another. FtsZ is found in almost all bacteria, many archaea, all chloroplasts and some mitochondria, where it is essential for cell division. FtsZ assembles the cytoskeletal scaffold of the Z ring that, along with additional proteins, constricts to divide the cell in two.
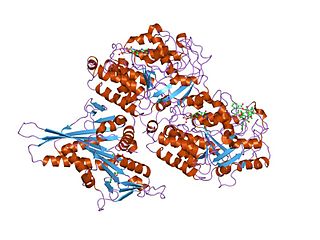
Tubulin in molecular biology can refer either to the tubulin protein superfamily of globular proteins, or one of the member proteins of that superfamily. α- and β-tubulins polymerize into microtubules, a major component of the eukaryotic cytoskeleton. Microtubules function in many essential cellular processes, including mitosis. Tubulin-binding drugs kill cancerous cells by inhibiting microtubule dynamics, which are required for DNA segregation and therefore cell division.

Tripartite motif-containing protein 5 also known as RING finger protein 88 is a protein that in humans is encoded by the TRIM5 gene. The alpha isoform of this protein, TRIM5α, is a retrovirus restriction factor, which mediates species-specific, early block to retrovirus infection.
SMC proteins represent a large family of ATPases that participate in many aspects of higher-order chromosome organization and dynamics. SMC stands for Structural Maintenance of Chromosomes.
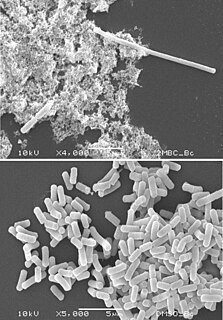
Filamentation is the anomalous growth of certain bacteria, such as Escherichia coli, in which cells continue to elongate but do not divide. The cells that result from elongation without division have multiple chromosomal copies. In the absence of antibiotics or other stressors, filamentation occurs at a low frequency in bacterial populations, the increased cell length protecting bacteria from protozoan predation and neutrophil phagocytosis by making ingestion of the cells more difficult. Filamentation is also a virulence factor thought to protect bacteria from antibiotics, and is associated with other aspects of bacterial virulence such as biofilm formation. The number and length of filaments within a bacterial population increases when the bacteria are treated with various chemical and physical agents. Some of the key genes involved in filamentation in E. coli include sulA and minCD.
Crescentin is a protein which is a bacterial relative of the intermediate filaments found in eukaryotic cells. Just as tubulins and actins, the other major cytoskeletal proteins, have prokaryotic homologs in, respectively, the FtsZ and MreB proteins, intermediate filaments are linked to the crescentin protein. Some of its homologs are erroneously labelled Chromosome segregation protein ParA. This protein family is found in Caulobacter and Methylobacterium.

The replisome is a complex molecular machine that carries out replication of DNA. The replisome first unwinds double stranded DNA into two single strands. For each of the resulting single strands, a new complementary sequence of DNA is synthesized. The net result is formation of two new double stranded DNA sequences that are exact copies of the original double stranded DNA sequence.
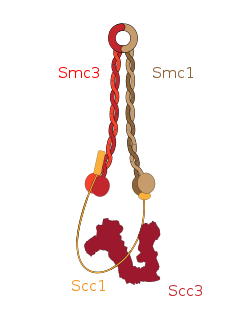
Cohesin is a protein complex that mediates sister chromatid cohesion, homologous recombination and DNA looping. Cohesin is formed of SMC3, SMC1, SCC1 and SCC3. Cohesin holds sister chromatids together after DNA replication until anaphase when removal of cohesin leads to separation of sister chromatids. The complex forms a ring-like structure and it is believed that sister chromatids are held together by entrapment inside the cohesin ring. Cohesin is a member of the SMC family of protein complexes which includes Condensin, MukBEF and SMC-ScpAB.
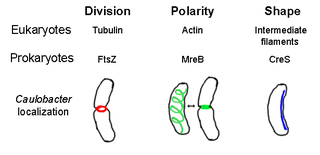
The prokaryotic cytoskeleton is the collective name for all structural filaments in prokaryotes. It was once thought that prokaryotic cells did not possess cytoskeletons, but advances in visualization technology and structure determination led to the discovery of filaments in these cells in the early 1990s. Not only have analogues for all major cytoskeletal proteins in eukaryotes been found in prokaryotes, cytoskeletal proteins with no known eukaryotic homologues have also been discovered. Cytoskeletal elements play essential roles in cell division, protection, shape determination, and polarity determination in various prokaryotes.
Fission, in biology, is the division of a single entity into two or more parts and the regeneration of those parts to separate entities resembling the original. The object experiencing fission is usually a cell, but the term may also refer to how organisms, bodies, populations, or species split into discrete parts. The fission may be binary fission, in which a single organism produces two parts, or multiple fission, in which a single entity produces multiple parts.

Tubulin/FtsZ family, GTPase domain is an evolutionary conserved protein domain.

The Min System is a mechanism composed of three proteins MinC, MinD, and MinE used by E. coli as a means of properly localizing the septum prior to cell division. Each component participates in generating a dynamic oscillation of FtsZ protein inhibition between the two bacterial poles to precisely specify the mid-zone of the cell, allowing the cell to accurately divide in two. This system is known to function in conjunction with a second negative regulatory system, the nucleoid occlusion system (NO), to ensure proper spatial and temporal regulation of chromosomal segregation and division.
The MinC protein is one of three proteins in the Min system encoded by the minB operon and which is required to generate pole to pole oscillations prior to bacterial cell division as a means of specifying the midzone of the cell. This function is achieved by preventing the formation of the divisome Z-ring around the poles.
The MinE protein is one of three proteins of the Min system encoded by the minB operon required to generate pole to pole oscillations prior to bacterial cell division as a means of specifying the midzone of the cell, as seen in E.coli.
Joe Lutkenhaus, Ph.D, is a Professor at the University of Kansas Medical Center. He received a B.S. in organic chemistry from Iowa state University and then a PhD in biochemistry for the University of California, Los Angeles. Following his PhD, Lutkenhaus pursued his postdoctoral studies with William Donachie at the University of Edinburgh and then continued at the University of Connecticut Health Science center. In 2002, Dr. Lutkenhaus became a fellow of the American Academy of Microbiology.

FtsA is a bacterial protein that is related to actin by overall structural similarity and in its ATP binding pocket.

In molecular biology, an actomyosin contractile ring is a prominent structure during cytokinesis. It forms perpendicular to the axis of the spindle apparatus towards the end of telophase, in which sister chromatids are identically separated at the opposite sides of the spindle forming nuclei. The actomyosin ring follows an orderly sequence of events: identification of the active division site, formation of the ring, constriction of the ring, and disassembly of the ring. It is composed of actin and myosin II bundles, thus the term actomyosin. The actomyosin ring operates in contractile motion, although the mechanism on how or what triggers the constriction is still an evolving topic. Other cytoskeletal proteins are also involved in maintaining stability of the ring. Apart from cytokinesis, in which the ring constricts as the cells divide, actomyosin ring constriction has also been found to activate during wound closure. During this process, actin filaments are degraded, preserving the thickness of the ring. After cytokinesis is complete, one of the two daughter cells inherits a remnant known as the midbody ring.

The divisome is a protein complex in bacteria that is responsible for cell division, constriction of inner and outer membranes during division, and peptidoglycan (PG) synthesis at the division site. The divisome is a membrane protein complex with proteins on both sides of the cytoplasmic membrane. In gram-negative cells it is located in the inner membrane. The divisome is nearly ubiquitous in bacteria although its composition may vary between species.













