Related Research Articles

A homeobox is a DNA sequence, around 180 base pairs long, found within genes that are involved in the regulation of patterns of anatomical development (morphogenesis) in animals, fungi, plants, and numerous single cell eukaryotes. Homeobox genes encode homeodomain protein products that are transcription factors sharing a characteristic protein fold structure that binds DNA to regulate expression of target genes. Homeodomain proteins regulate gene expression and cell differentiation during early embryonic development, thus mutations in homeobox genes can cause developmental disorders.
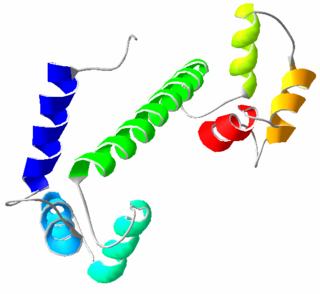
Calmodulin (CaM) (an abbreviation for calcium-modulated protein) is a multifunctional intermediate calcium-binding messenger protein expressed in all eukaryotic cells. It is an intracellular target of the secondary messenger Ca2+, and the binding of Ca2+ is required for the activation of calmodulin. Once bound to Ca2+, calmodulin acts as part of a calcium signal transduction pathway by modifying its interactions with various target proteins such as kinases or phosphatases.
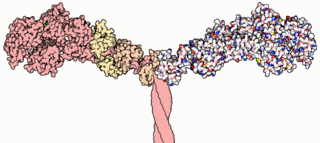
Myosins are a superfamily of motor proteins best known for their roles in muscle contraction and in a wide range of other motility processes in eukaryotes. They are ATP-dependent and responsible for actin-based motility. The term was originally used to describe a group of similar ATPases found in the cells of both striated muscle tissue and smooth muscle tissue. Following the discovery by Pollard and Korn (1973) of enzymes with myosin-like function in Acanthamoeba castellanii, a global range of divergent myosin genes have been discovered throughout the realm of eukaryotes.
A histone fold is a structurally conserved motif found near the C-terminus in every core histone sequence in a histone octamer responsible for the binding of histones into heterodimers.

Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers – specifically polypeptides – formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a residue indicating a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their three-dimensional structure. This is the topic of the scientific field of structural biology, which employs techniques such as X-ray crystallography, NMR spectroscopy, cryo electron microscopy (cryo-EM) and dual polarisation interferometry to determine the structure of proteins.

Titin, also known as connectin, is a protein that in humans is encoded by the TTN gene. Titin is a giant protein, greater than 1 µm in length, that functions as a molecular spring which is responsible for the passive elasticity of muscle. It comprises 244 individually folded protein domains connected by unstructured peptide sequences. These domains unfold when the protein is stretched and refold when the tension is removed.

Motor proteins are a class of molecular motors that can move along the cytoplasm of animal cells. They convert chemical energy into mechanical work by the hydrolysis of ATP. Flagellar rotation, however, is powered by a proton pump.

Myosin-9 also known as myosin, heavy chain 9, non-muscle or non-muscle myosin heavy chain IIa (NMMHC-IIA) is a protein which in humans is encoded by the MYH9 gene.
In enzymology, a myosin-heavy-chain kinase is an enzyme that catalyzes the chemical reaction

Myosin-10 also known as myosin heavy chain 10 or non-muscle myosin IIB (NM-IIB) is a protein that in humans is encoded by the MYH10 gene. Non-muscle myosins are expressed in a wide variety of tissues, but NM-IIB is the only non-muscle myosin II isoform expressed in cardiac muscle, where it localizes to adherens junctions within intercalated discs. NM-IIB is essential for normal development of cardiac muscle and for integrity of intercalated discs. Mutations in MYH10 have been identified in patients with left atrial enlargement.

Citron Rho-interacting kinase is an enzyme that in humans is encoded by the CIT gene.

Myosin X, also known as MYO10, is a protein that in humans is encoded by the MYO10 gene.

Myosin-1, also known as 'striated muscle myosin heavy chain 1', is a protein that in humans is encoded by the MYH1 gene. This gene is most highly expressed in fast type IIX/D muscle fibres of vertebrates and encodes a protein found uniquely in striated muscle; it is a class II myosin with a long coiled coil tail that dimerizes and should not be confused with 'Myosin 1' encoded by the MYO1 family of genes (MYO1A-MYO1H). Class I MYO1 genes function in many cell types throughout biology and are single-headed membrane-binding myosins that lack a long coiled coil tail.

Myosin-Ia is a protein that in humans is encoded by the MYO1A gene.

Protein misato homolog 1 is a protein that in humans is encoded by the MSTO1 gene.

The insulin/IGF/relaxin family is a group of evolutionary related proteins which possess a variety of hormonal activities. Family members in human include two subfamilies:
The Walker A and Walker B motifs are protein sequence motifs, known to have highly conserved three-dimensional structures. These were first reported in ATP-binding proteins by Walker and co-workers in 1982.

The rhomboid proteases are a family of enzymes that exist in almost all species. They are proteases: they cut the polypeptide chain of other proteins. This proteolytic cleavage is irreversible in cells, and an important type of cellular regulation. Although proteases are one of the earliest and best studied class of enzyme, rhomboids belong to a much more recently discovered type: the intramembrane proteases. What is unique about intramembrane proteases is that their active sites are buried in the lipid bilayer of cell membranes, and they cleave other transmembrane proteins within their transmembrane domains. About 30% of all proteins have transmembrane domains, and their regulated processing often has major biological consequences. Accordingly, rhomboids regulate many important cellular processes, and may be involved in a wide range of human diseases.

The myosin head is the part of the thick myofilament made up of myosin that acts in muscle contraction, by sliding over thin myofilaments of actin. Myosin is the major component of the thick filaments and most myosin molecules are composed of a head, neck, and tail domain; the myosin head binds to thin filamentous actin, and uses ATP hydrolysis to generate force and "walk" along the thin filament. Myosin exists as a hexamer of two heavy chains, two alkali light chains, and two regulatory light chains. The heavy chain can be subdivided into the globular head at the N-terminal and the coiled-coil rod-like tail at the C-terminal, although some forms have a globular region in their C-terminal.

In molecular biology, the GCM transcription factors are a family of proteins which contain a GCM motif. The GCM motif is a domain that has been identified in proteins belonging to a family of transcriptional regulators involved in fundamental developmental processes which comprise Drosophila melanogaster GCM and its mammalian homologues. In GCM transcription factors the N-terminal moiety contains a DNA-binding domain of 150 amino acids. Sequence conservation is highest in this GCM domain. In contrast, the C-terminal moiety contains one or two transactivating regions and is only poorly conserved.
References
- ↑ Kimura M, Okano Y (April 2007). "Human Misato regulates mitochondrial distribution and morphology". Exp. Cell Res. 313 (7): 1393–404. doi:10.1016/j.yexcr.2007.02.004. PMID 17349998.
- ↑ Miklos GL, Yamamoto M, Burns RG, Maleszka R (May 1997). "An essential cell division gene of Drosophila, absent from Saccharomyces, encodes an unusual protein with tubulin-like and myosin-like peptide motifs". Proc. Natl. Acad. Sci. U.S.A. 94 (10): 5189–94. Bibcode:1997PNAS...94.5189M. doi: 10.1073/pnas.94.10.5189 . PMC 24654 . PMID 9144213.