
Chlamydomonas reinhardtii is a single-cell green alga about 10 micrometres in diameter that swims with two flagella. It has a cell wall made of hydroxyproline-rich glycoproteins, a large cup-shaped chloroplast, a large pyrenoid, and an eyespot that senses light.
The transcriptome is the set of all RNA transcripts, including coding and non-coding, in an individual or a population of cells. The term can also sometimes be used to refer to all RNAs, or just mRNA, depending on the particular experiment. The term transcriptome is a portmanteau of the words transcript and genome; it is associated with the process of transcript production during the biological process of transcription.

Phaeodactylum tricornutum is a diatom. It is the only species in the genus Phaeodactylum. Unlike other diatoms, P. tricornutum can exist in different morphotypes and changes in cell shape can be stimulated by environmental conditions. This feature can be used to explore the molecular basis of cell shape control and morphogenesis. Unlike most diatoms, P. tricornutum can grow in the absence of silicon and can survive without making silicified frustules. This provides opportunities for experimental exploration of silicon-based nanofabrication in diatoms.
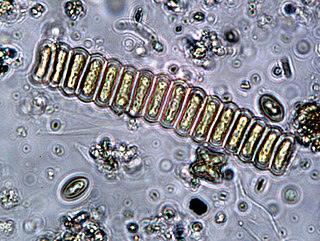
Scenedesmus is a genus of green algae, in the class Chlorophyceae. They are colonial and non-motile. They are one of the most common components of phytoplankton in freshwater habitats worldwide.

Selenastraceae is a family of green algae in the order Sphaeropleales. Members of this family are common components of the phytoplankton in freshwater habitats worldwide. A few species have been found in brackish and marine habitats, such as in the Baltic Sea.
Auxenochlorella protothecoides, formerly known as Chlorella protothecoides, is a facultative heterotrophic green alga in the family Chlorellaceae. It is known for its potential application in biofuel production. It was first characterized as a distinct algal species in 1965, and has since been regarded as a separate genus from Chlorella due its need for thiamine for growth. Auxenochlorella species have been found in a wide variety of environments from acidic volcanic soil in Italy to the sap of poplar trees in the forests of Germany. Its use in industrial processes has been studied, as the high lipid content of the alga during heterotrophic growth is promising for biodiesel; its use in wastewater treatment has been investigated, as well.

Choricystis is a genus of green algae in the class Trebouxiophyceae, considered a characteristic picophytoplankton in freshwater ecosystems. Choricystis, especially the type species Choricystis minor, has been proposed as an effective source of fatty acids for biofuels. Choricystis algacultures have been shown to survive on wastewater. In particular, Choricystis has been proposed as a biological water treatment system for industrial waste produced by the processing of dairy goods.

Algae fuel, algal biofuel, or algal oil is an alternative to liquid fossil fuels that uses algae as its source of energy-rich oils. Also, algae fuels are an alternative to commonly known biofuel sources, such as corn and sugarcane. When made from seaweed (macroalgae) it can be known as seaweed fuel or seaweed oil.

Heat shock factor protein 4 is a protein that in humans is encoded by the HSF4 gene.

RNA-Seq is a technique that uses next-generation sequencing to reveal the presence and quantity of RNA molecules in a biological sample, providing a snapshot of gene expression in the sample, also known as transcriptome.

Nannochloropsis is a genus of algae comprising six known species. The genus in the current taxonomic classification was first termed by Hibberd (1981). The species have mostly been known from the marine environment but also occur in fresh and brackish water. All of the species are small, nonmotile spheres which do not express any distinct morphological features that can be distinguished by either light or electron microscopy. The characterisation is mostly done by rbcL gene and 18S rRNA sequence analysis.
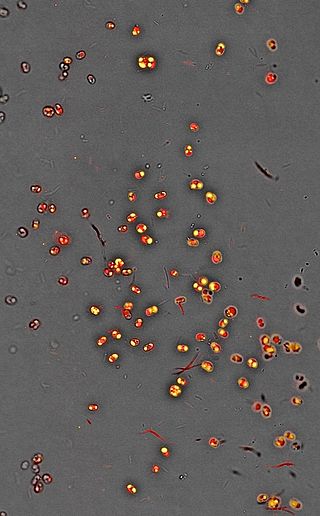
Nannochloropsis is a genus of alga within the heterokont line of eukaryotes, that is being investigated for biofuel production. One marine Nannochloropsis species has been shown to be suitable for algal biofuel production due to its ease of growth and high oil content, mainly unsaturated fatty acids and a significant percentage of palmitic acid. It also contains enough unsaturated fatty acid linolenic acid and polyunsaturated acid for a quality biodiesel.
Schizochytrium is a genus of unicellular eukaryotes in the family Thraustochytriaceae, which are found in coastal marine habitats. They are assigned to the Stramenopiles (heterokonts), a group which also contains kelp and various microalgae.

Tetradesmus obliquus is a green algae species of the family Scenedesmaceae. It is commonly known by its synonym, Scenedesmus obliquus. It is a common species found in a variety of freshwater habitats.
Sammy Boussiba is a professor emeritus at the French Associates Institute for Agriculture and Biotechnology of Drylands at the Jacob Blaustein Institutes for Desert Research at Ben-Gurion University of the Negev, Israel.

Chlorella vulgaris is a species of green microalga in the division Chlorophyta. It is mainly used as a dietary supplement or protein-rich food additive in Japan.
Christoph Benning is a German–American plant biologist. He is an MSU Foundation Professor and University Distinguished Professor at Michigan State University. Benning's research into lipid metabolism in plants, algae and photosynthetic bacteria, led him to be named Editor-in-Chief of The Plant Journal in October 2008.
An oleaginous microorganism is a type of microbe that accumulates lipid as a normal part of its metabolism. Oleaginous microbes may accumulate an array of different lipid compounds, including polyhydroxyalkanoates, triacylglycerols, and wax esters. Various microorganisms, including bacteria, fungi, and yeast, are known to accumulate lipids. These organisms are often researched for their potential use in producing fuels from waste products.










