
Oxidative phosphorylation or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in order to produce adenosine triphosphate (ATP). In eukaryotes, this takes place inside mitochondria. Almost all aerobic organisms carry out oxidative phosphorylation. This pathway is so pervasive because it releases more energy than alternative fermentation processes such as anaerobic glycolysis.

An electron transport chain (ETC) is a series of protein complexes and other molecules that transfer electrons from electron donors to electron acceptors via redox reactions (both reduction and oxidation occurring simultaneously) and couples this electron transfer with the transfer of protons (H+ ions) across a membrane. Many of the enzymes in the electron transport chain are membrane-bound.
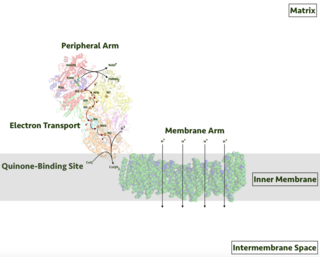
Respiratory complex I, EC 7.1.1.2 is the first large protein complex of the respiratory chains of many organisms from bacteria to humans. It catalyzes the transfer of electrons from NADH to coenzyme Q10 (CoQ10) and translocates protons across the inner mitochondrial membrane in eukaryotes or the plasma membrane of bacteria.
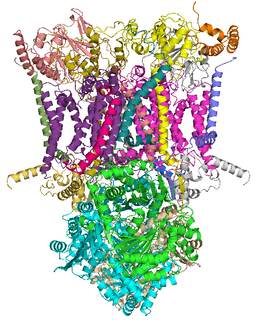
The coenzyme Q : cytochrome c – oxidoreductase, sometimes called the cytochrome bc1 complex, and at other times complex III, is the third complex in the electron transport chain, playing a critical role in biochemical generation of ATP. Complex III is a multisubunit transmembrane protein encoded by both the mitochondrial and the nuclear genomes. Complex III is present in the mitochondria of all animals and all aerobic eukaryotes and the inner membranes of most eubacteria. Mutations in Complex III cause exercise intolerance as well as multisystem disorders. The bc1 complex contains 11 subunits, 3 respiratory subunits, 2 core proteins and 6 low-molecular weight proteins.

Succinate dehydrogenase (SDH) or succinate-coenzyme Q reductase (SQR) or respiratory complex II is an enzyme complex, found in many bacterial cells and in the inner mitochondrial membrane of eukaryotes. It is the only enzyme that participates in both the citric acid cycle and the electron transport chain. Histochemical analysis showing high succinate dehydrogenase in muscle demonstrates high mitochondrial content and high oxidative potential.

In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.

Glycerol-3-phosphate dehydrogenase (GPDH) is an enzyme that catalyzes the reversible redox conversion of dihydroxyacetone phosphate to sn-glycerol 3-phosphate.

Electron-transferring-flavoprotein dehydrogenase is an enzyme that transfers electrons from electron-transferring flavoprotein in the mitochondrial matrix, to the ubiquinone pool in the inner mitochondrial membrane. It is part of the electron transport chain. The enzyme is found in both prokaryotes and eukaryotes and contains a flavin and FE-S cluster. In humans, it is encoded by the ETFDH gene. Deficiency in ETF dehydrogenase causes the human genetic disease multiple acyl-CoA dehydrogenase deficiency.

In enzymology, a NADH dehydrogenase (quinone) (EC 1.6.5.11) is an enzyme that catalyzes the chemical reaction
In enzymology, a NAD(P)H dehydrogenase (quinone) (EC 1.6.5.2) is an enzyme that catalyzes the chemical reaction
In enzymology, a NADPH dehydrogenase (quinone) (EC 1.6.5.10) is an enzyme that catalyzes the chemical reaction

NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 6, also known as complex I-B17, is a protein that in humans is encoded by the NDUFB6 gene. NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 6, is an accessory subunit of the NADH dehydrogenase (ubiquinone) complex, located in the mitochondrial inner membrane. It is also known as Complex I and is the largest of the five complexes of the electron transport chain.

FAD-dependent oxidoreductase domain-containing protein 1 (FOXRED1), also known as H17, or FP634 is an enzyme that in humans is encoded by the FOXRED1 gene. FOXRED1 is an oxidoreductase and complex I-specific molecular chaperone involved in the assembly and stabilization of NADH dehydrogenase (ubiquinone) also known as complex I, which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Mutations in FOXRED1 have been associated with Leigh syndrome and infantile-onset mitochondrial encephalopathy.

NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1 is an enzyme that in humans is encoded by the NDUFB1 gene. NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa is an accessory subunit of the NADH dehydrogenase (ubiquinone) complex, located in the mitochondrial inner membrane. It is also known as Complex I and is the largest of the five complexes of the electron transport chain.

NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7, also known as complex I-B18, is an enzyme that in humans is encoded by the NDUFB7 gene. NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 7 is an accessory subunit of the NADH dehydrogenase (ubiquinone) complex, located in the mitochondrial inner membrane. It is also known as Complex I and is the largest of the five complexes of the electron transport chain.

NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 is an enzyme that in humans is encoded by the NDUFB10 gene. NADH dehydrogenase (ubiquinone) 1 beta subcomplex subunit 10 is an accessory subunit of the NADH dehydrogenase (ubiquinone) complex, located in the mitochondrial inner membrane. It is also known as Complex I and is the largest of the five complexes of the electron transport chain.

Class 2 dihydroorotate dehydrogenases is an enzyme with systematic name (S)-dihydroorotate:quinone oxidoreductase. This enzyme catalyses the electron transfer from dihydroorotate to a quinone :

Fumarate reductase (quinol) is an enzyme with systematic name succinate:quinone oxidoreductase. This enzyme catalyzes the following chemical reaction:
The Monovalent Cation (K+ or Na+):Proton Antiporter-3 (CPA3) Family (TC# 2.A.63) is a member of the Na+ transporting Mrp superfamily. The CPA3 family consists of bacterial multicomponent K+:H+ and Na+:H+ antiporters. The best characterized systems are the PhaABCDEFG system of Sinorhizobium meliloti (TC# 2.A.63.1.1) that functions in pH adaptation and as a K+ efflux system, and the MnhABCDEFG system of Staphylococcus aureus (TC# 2.A.63.1.3) that functions as a Na+ efflux Na+:H+ antiporter.
The H+-translocating F420H2 Dehydrogenase (F420H2DH) Family(TC# 3.D.9) is a member of the Na+ transporting Mrp superfamily. A single F420H2 dehydrogenase (also referred to as F420H2:quinol oxidoreductase) from the methanogenic archaeon, Methanosarcina mazei Gö1, has been shown to be a redox driven proton pump. The F420H2DH of M. mazei has a molecular size of about 120 kDa and contains Fe-S clusters and FAD. A similar five-subunit enzyme has been isolated from Methanolobus tindarius. The sulfate-reducing Archaeoglobus fulgidus (and several other archaea) also have this enzyme.


















