
In biochemistry, the DNA methyltransferase family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor.

Mercaptopurine (6-MP), sold under the brand name Purinethol among others, is a medication used for cancer and autoimmune diseases. Specifically it is used to treat acute lymphocytic leukemia (ALL), acute promyelocytic leukemia (APL), Crohn's disease, and ulcerative colitis. For acute lymphocytic leukemia it is generally used with methotrexate. It is taken by mouth.

Thiopurine methyltransferase or thiopurine S-methyltransferase (TPMT) is an enzyme that in humans is encoded by the TPMT gene. A pseudogene for this locus is located on chromosome 18q.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.
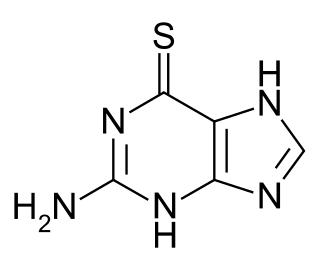
Tioguanine, also known as thioguanine or 6-thioguanine (6-TG) is a medication used to treat acute myeloid leukemia (AML), acute lymphocytic leukemia (ALL), and chronic myeloid leukemia (CML). Long-term use is not recommended. It is given by mouth.

Histamine N-methyltransferase (HNMT) is a cytoplasmic protein encoded by the HNMT gene in humans. It belongs to the methyltransferases superfamily of enzymes and plays a crucial role in the inactivation of histamine, a biogenic amine involved in various physiological processes. Methyltransferases are present in every life form including archaeans, with 230 families of methyltransferases found across species.

Phosphatidylethanolamine N-methyltransferase is a transferase enzyme which converts phosphatidylethanolamine (PE) to phosphatidylcholine (PC) in the liver. In humans it is encoded by the PEMT gene within the Smith–Magenis syndrome region on chromosome 17.

Enhancer of zeste homolog 2 (EZH2) is a histone-lysine N-methyltransferase enzyme encoded by EZH2 gene, that participates in histone methylation and, ultimately, transcriptional repression. EZH2 catalyzes the addition of methyl groups to histone H3 at lysine 27, by using the cofactor S-adenosyl-L-methionine. Methylation activity of EZH2 facilitates heterochromatin formation thereby silences gene function. Remodeling of chromosomal heterochromatin by EZH2 is also required during cell mitosis.
Protein arginine N-methyltransferase 1 is an enzyme that in humans is encoded by the PRMT1 gene. The HRMT1L2 gene encodes a protein arginine methyltransferase that functions as a histone methyltransferase specific for histone H4.

DNA (cytosine-5)-methyltransferase 3A (DNMT3A) is an enzyme that catalyzes the transfer of methyl groups to specific CpG structures in DNA, a process called DNA methylation. The enzyme is encoded in humans by the DNMT3A gene.

Nicotinamide phosphoribosyltransferase, formerly known as pre-B-cell colony-enhancing factor 1 (PBEF1) or visfatin for its extracellular form (eNAMPT), is an enzyme that in humans is encoded by the NAMPT gene. The intracellular form of this protein (iNAMPT) is the rate-limiting enzyme in the nicotinamide adenine dinucleotide (NAD+) salvage pathway that converts nicotinamide to nicotinamide mononucleotide (NMN) which is responsible for most of the NAD+ formation in mammals. iNAMPT can also catalyze the synthesis of NMN from phosphoribosyl pyrophosphate (PRPP) when ATP is present. eNAMPT has been reported to be a cytokine (PBEF) that activates TLR4, that promotes B cell maturation, and that inhibits neutrophil apoptosis.

Sulfotransferase 1A1 is an enzyme that in humans is encoded by the SULT1A1 gene.

Estrogen sulfotransferase is an enzyme that in humans is encoded by the SULT1E1 gene.

UDP-glucuronosyltransferase 2B15 is an enzyme that in humans is encoded by the UGT2B15 gene.

Sulfotransferase 1A2 is an enzyme that in humans is encoded by the SULT1A2 gene.

Glycine N-methyltransferase is an enzyme that in humans is encoded by the GNMT gene.

1-Methylnicotinamide (trigonellamide) is a prototypic organic cation. 1-Methylnicotinamide is the methylated amide of Nicotinamide (niacinamide, vitamin B3).

Arsenite methyltransferase is an enzyme that in humans is encoded by the AS3MT gene.

Bile salt sulfotransferase also known as hydroxysteroid sulfotransferase (HST) or sulfotransferase 2A1 (ST2A1) is an enzyme that in humans is encoded by the SULT2A1 gene.

Cancer epigenetics is the study of epigenetic modifications to the DNA of cancer cells that do not involve a change in the nucleotide sequence, but instead involve a change in the way the genetic code is expressed. Epigenetic mechanisms are necessary to maintain normal sequences of tissue specific gene expression and are crucial for normal development. They may be just as important, if not even more important, than genetic mutations in a cell's transformation to cancer. The disturbance of epigenetic processes in cancers, can lead to a loss of expression of genes that occurs about 10 times more frequently by transcription silencing than by mutations. As Vogelstein et al. points out, in a colorectal cancer there are usually about 3 to 6 driver mutations and 33 to 66 hitchhiker or passenger mutations. However, in colon tumors compared to adjacent normal-appearing colonic mucosa, there are about 600 to 800 heavily methylated CpG islands in the promoters of genes in the tumors while these CpG islands are not methylated in the adjacent mucosa. Manipulation of epigenetic alterations holds great promise for cancer prevention, detection, and therapy. In different types of cancer, a variety of epigenetic mechanisms can be perturbed, such as the silencing of tumor suppressor genes and activation of oncogenes by altered CpG island methylation patterns, histone modifications, and dysregulation of DNA binding proteins. There are several medications which have epigenetic impact, that are now used in a number of these diseases.



























