
The aspS RNA motif is a conserved RNA structure that was discovered by bioinformatics. aspS motifs are found in a specific lineage of Actinomycetota.

The COG2908 RNA motif is a conserved RNA structure that was discovered by bioinformatics. COG2908 motif RNAs are found in genomic sequences extracted from fresh water environments. They have not, as of 2018, been detected in any classified organism.

The COG3943 RNA motif is a conserved RNA structure that was discovered by bioinformatics. COG3943 motifs are found in unknown bacteria whose genomic DNA was isolated from cow rumen. As of 2018, there is no specific, classified organism that is known to contain a COG3943 motif RNA.
The DUF3085 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF3085 motifs are found in one species in the genus Thauera, as well as various metagenomic sequences obtained from environmental DNA.
The gltS RNA motif is a conserved RNA structure that was discovered by bioinformatics. gltS motifs are found in the bacterial lineage Vibrionaceae.

The hya RNA motif is a conserved RNA structure that was discovered by bioinformatics. hya motif RNAs are found in Actinomycetota.
The ldcC RNA motif is a conserved RNA structure that was discovered by bioinformatics. ldcC motif RNAs are found in Bacillota and two species of Spirochaetota.
The leuA-Halobacteria RNA motif is a conserved RNA structure that was discovered by bioinformatics. leuA-Halobacteria motifs are found in Halobacteriaceae, a lineage of archaea.
lysM RNA motifs are conserved RNA structures that were discovered by bioinformatics. Such bacterial motifs are defined by consistently being upstream of 'lysM' genes, which encode lysin protein domains, a conserved domain that participates in cell wall degradation. lysM motif RNAs likely function as cis-regulatory elements, in view of their positions upstream of protein-coding genes, although this hypothesis is not certain.
malK RNA motifs are conserved RNA structures that were discovered by bioinformatics. They are defined by being consistently located upstream of malK genes, which encode an ATPase that is used by transporters whose ligand is likely a kind of sugar. Most of these genes are annotated either as transporting maltose or glycerol-3-phosphate, however the substrate of the transporters associated with malK motif RNAs has not been experimentally determined. All known types of malK RNA motif are generally located nearby to the Shine-Dalgarno sequence of the downstream gene.

The nhaA-I RNA motif is a conserved RNA structure that was discovered by bioinformatics. nhaA-I motif RNAs are found in Acidobacteriota, alpha-, beta- and Gammaproteobacteria, Verrucomicrobiota and the tentative phylum NC10.

The NLPC-P60 RNA motif is a conserved RNA structure that was discovered by bioinformatics. NLPC-P60 motif RNAs are found in Streptomyces.
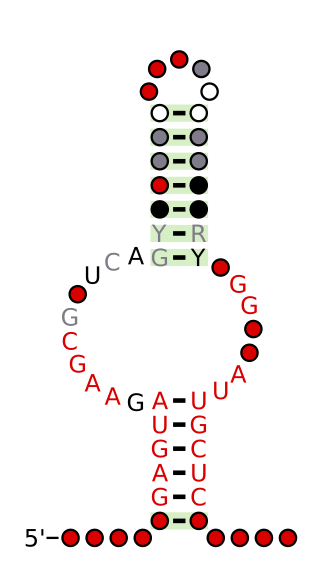
The NMT1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. NMT1 motif RNAs are found in Pseudomonadota. There is also one NMT1 RNA in each of Bacteroidota and Actinomycetota, but these appear to be the result of recent horizontal gene transfer or sequence contamination before or during genome sequencing
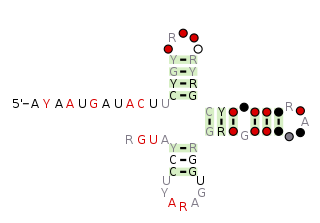
The pemK RNA motif is a conserved RNA structure that was discovered by bioinformatics. pemK motif RNAs are found in organisms within the phylum Bacillota, and is very widespread in this phylum.

The PGK RNA motif is a conserved RNA structure that was discovered by bioinformatics. PGK motif RNAs are found in metagenomic sequences isolated from the gastrointestinal tract of mammals. PGK RNAs have not yet been detected in a classified organism.
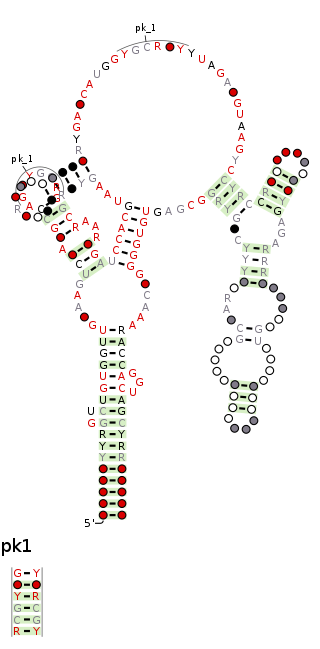
The raiA RNA motif is a conserved RNA structure that was discovered by bioinformatics. raiA motif RNAs are found in Actinomycetota and Bacillota, and have many conserved features—including conserved nucleotide positions, conserved secondary structures and associated protein-coding genes—in both of these phyla. Some conserved features of the raiA RNA motif suggest that they function as cis-regulatory elements, but other aspects of the motif suggest otherwise.

The Rhodo-rpoB RNA motif is a conserved RNA structure that was discovered by bioinformatics. Rhodo-rpoB motifs are found in Rhodobacterales.

The Rothia-sucC RNA motif is a conserved RNA structure that was discovered by bioinformatics. Rothia-sucC motif RNAs are found in the actinobacterial genus Rothia.

The sul1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Energetically stable tetraloops often occur in this motif. sul1 motif RNAs are found in Alphaproteobacteria.

The uup RNA motif is a conserved RNA structure that was discovered by bioinformatics. uup motif RNAs are found in Bacillota and Gammaproteobacteria.














