Ferredoxins are iron–sulfur proteins that mediate electron transfer in a range of metabolic reactions. The term "ferredoxin" was coined by D.C. Wharton of the DuPont Co. and applied to the "iron protein" first purified in 1962 by Mortenson, Valentine, and Carnahan from the anaerobic bacterium Clostridium pasteurianum.

Sulfate-reducing microorganisms (SRM) or sulfate-reducing prokaryotes (SRP) are a group composed of sulfate-reducing bacteria (SRB) and sulfate-reducing archaea (SRA), both of which can perform anaerobic respiration utilizing sulfate (SO2−
4) as terminal electron acceptor, reducing it to hydrogen sulfide (H2S). Therefore, these sulfidogenic microorganisms "breathe" sulfate rather than molecular oxygen (O2), which is the terminal electron acceptor reduced to water (H2O) in aerobic respiration.
Nitrite reductase refers to any of several classes of enzymes that catalyze the reduction of nitrite. There are two classes of NIR's. A multi haem enzyme reduces NO2− to a variety of products. Copper containing enzymes carry out a single electron transfer to produce nitric oxide.
Microbial metabolism is the means by which a microbe obtains the energy and nutrients it needs to live and reproduce. Microbes use many different types of metabolic strategies and species can often be differentiated from each other based on metabolic characteristics. The specific metabolic properties of a microbe are the major factors in determining that microbe's ecological niche, and often allow for that microbe to be useful in industrial processes or responsible for biogeochemical cycles.
In enzymology, a sulfhydrogenase, also known as sulfur reductase, is an enzyme that catalyzes the reduction of elemental sulfur or polysulfide to hydrogen sulfide using hydrogen as electron donor.
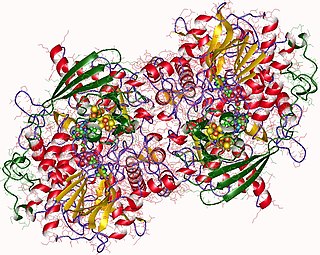
Adenylyl-sulfate reductase is an enzyme that catalyzes the chemical reaction of the reduction of adenylyl-sulfate/adenosine-5'-phosphosulfate (APS) to sulfite through the use of an electron donor cofactor. The products of the reaction are AMP and sulfite, as well as an oxidized electron donor cofactor.
Adenylyl-sulfate reductase (glutathione) is an enzyme that catalyzes the chemical reaction
Adenylyl-sulfate reductase (thioredoxin) is an enzyme that catalyzes the chemical reaction
In enzymology, a ferredoxin—nitrate reductase (EC 1.7.7.2) is an enzyme that catalyzes the chemical reaction
In enzymology, a hydrogensulfite reductase (EC 1.8.99.3) is an enzyme that catalyzes the chemical reaction
In enzymology, a nitrite reductase [NAD(P)H] (EC 1.7.1.4) is an enzyme that catalyzes the chemical reaction
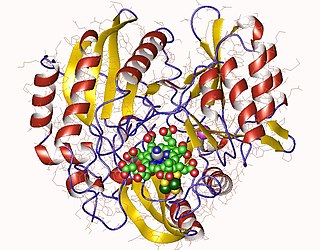
Sulfite reductase (NADPH) (EC 1.8.1.2, sulfite (reduced nicotinamide adenine dinucleotide phosphate) reductase, NADPH-sulfite reductase, NADPH-dependent sulfite reductase, H2S-NADP oxidoreductase, sulfite reductase (NADPH2)) is an enzyme with systematic name hydrogen-sulfide:NADP+ oxidoreductase. This enzyme catalises the following chemical reaction
Sulfur is metabolized by all organisms, from bacteria and archaea to plants and animals. Sulfur can have an oxidation state from -2 to +6 and is reduced or oxidized by a diverse range of organisms. The element is present in proteins, sulfate esters of polysaccharides, steroids, phenols, and sulfur-containing coenzymes.
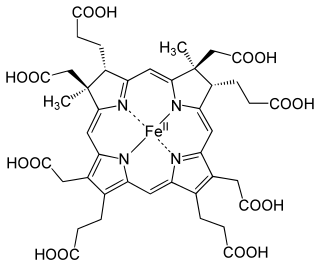
Siroheme is a heme-like prosthetic group at the active sites of some enzymes to accomplish the six-electron reduction of sulfur and nitrogen. It is a cofactor at the active site of sulfite reductase, which plays a major role in sulfur assimilation pathway, converting sulfite into sulfide, which can be incorporated into the organic compound homocysteine.

Short-chain acyl-CoA dehydrogenase is an enzyme with systematic name short-chain acyl-CoA:electron-transfer flavoprotein 2,3-oxidoreductase. This enzyme catalyses the following chemical reaction
Sulfide:quinone reductase is an enzyme with systematic name sulfide:quinone oxidoreductase. This enzyme catalyses the following chemical reaction

Dissimilatory sulfate reduction is a form of anaerobic respiration that uses sulfate as the terminal electron acceptor to produce hydrogen sulfide. This metabolism is found in some types of bacteria and archaea which are often termed sulfate-reducing organisms. The term "dissimilatory" is used when hydrogen sulfide is produced in an anaerobic respiration process. By contrast, the term "assimilatory" would be used in relation to the biosynthesis of organosulfur compounds, even though hydrogen sulfide may be an intermediate.

In enzymology, an aldehyde ferredoxin oxidoreductase (EC 1.2.7.5) is an enzyme that catalyzes the chemical reaction

Microbial oxidation of sulfur is the oxidation of sulfur by microorganisms to build their structural components. The oxidation of inorganic compounds is the strategy primarily used by chemolithotrophic microorganisms to obtain energy to survive, grow and reproduce. Some inorganic forms of reduced sulfur, mainly sulfide (H2S/HS−) and elemental sulfur (S0), can be oxidized by chemolithotrophic sulfur-oxidizing prokaryotes, usually coupled to the reduction of oxygen (O2) or nitrate (NO3−). Anaerobic sulfur oxidizers include photolithoautotrophs that obtain their energy from sunlight, hydrogen from sulfide, and carbon from carbon dioxide (CO2).
Dissimilatory sulfite reductase is an enzyme that participates in sulfur metabolism in dissimilatory sulfate reduction.









