Calcium release-activated channels (CRAC) are specialized plasma membrane Ca2+ ion channels. When calcium ions (Ca2+) are depleted from the endoplasmic reticulum (a major store of Ca2+) of mammalian cells, the CRAC channel is activated to slowly replenish the level of calcium in the endoplasmic reticulum. The Ca2+ Release-activated Ca2+ (CRAC) Channel (CRAC-C) Family (TC# 1.A.52) is a member of the Cation Diffusion Facilitator (CDF) Superfamily. These proteins typically have between 4 and 6 transmembrane α-helical spanners (TMSs). The 4 TMS CRAC channels arose by loss of 2TMSs from 6TMS CDF carriers, an example of 'reverse' evolution'.
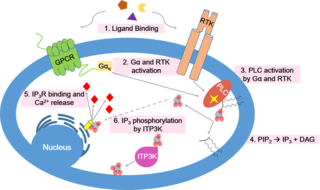
Calcium signaling is the use of calcium ions (Ca2+) to communicate and drive intracellular processes often as a step in signal transduction. Ca2+ is important for cellular signalling, for once it enters the cytosol of the cytoplasm it exerts allosteric regulatory effects on many enzymes and proteins. Ca2+ can act in signal transduction resulting from activation of ion channels or as a second messenger caused by indirect signal transduction pathways such as G protein-coupled receptors.

Calnexin (CNX) is a 67kDa integral protein (that appears variously as a 90kDa, 80kDa, or 75kDa band on western blotting depending on the source of the antibody) of the endoplasmic reticulum (ER). It consists of a large (50 kDa) N-terminal calcium-binding lumenal domain, a single transmembrane helix and a short (90 residues), acidic cytoplasmic tail.
Store-operated channels (SOCs) are ion channels located in the plasma membrane of cells. These channels are most studied in regard to their role in calcium entry into the cytoplasm from extracellular milieu. There are other SOC channels selective to other ions. Calcium SOCs are especially important for the cell because they are the major source of intracellular calcium; and calcium itself is involved in a wide array of vital cellular functions. SOCs are so called because they are activated by intracellular calcium stores depletion by both physiological or pharmacological processes.

Stromal interaction molecule 1 is a protein that in humans is encoded by the STIM1 gene. STIM1 has a single transmembrane domain, and is localized to the endoplasmic reticulum, and to a lesser extent to the plasma membrane.
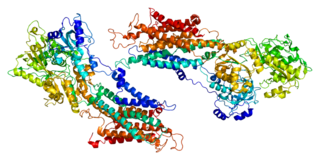
Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (SERCA1) also known as Calcium pump 1, is an enzyme that in humans is encoded by the ATP2A1 gene.

Eukaryotic translation initiation factor 2-alpha kinase 3, also known as protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK), is an enzyme that in humans is encoded by the EIF2AK3 gene.

Cav1.1 also known as the calcium channel, voltage-dependent, L type, alpha 1S subunit, (CACNA1S), is a protein which in humans is encoded by the CACNA1S gene. It is also known as CACNL1A3 and the dihydropyridine receptor.

Ceroid-lipofuscinosis neuronal protein 6 is a protein that in humans is encoded by the CLN6 gene.

KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1, also known as KDELR1, is a protein which in humans is encoded by the KDELR1 gene.

Protein transport protein Sec61 subunit gamma is a protein that in humans is encoded by the SEC61G gene.

Stromal interaction molecule 2 (STIM2) is a protein that in humans is encoded by the STIM2 gene.

SARAF is a protein that in humans is encoded by the SARAF gene, formerly known as TMEM66.

Protein orai-2 is a protein that in humans is encoded by the ORAI2 gene.
Membrane contact sites (MCS) are close appositions between two organelles. Ultrastructural studies typically reveal an intermembrane distance in the order of the size of a single protein, as small as 10 nm or wider, with no clear upper limit. These zones of apposition are highly conserved in evolution. These sites are thought to be important to facilitate signalling, and they promote the passage of small molecules, including ions, lipids and reactive oxygen species. MCS are important in the function of the endoplasmic reticulum (ER), since this is the major site of lipid synthesis within cells. The ER makes close contact with many organelles, including mitochondria, Golgi, endosomes, lysosomes, peroxisomes, chloroplasts and the plasma membrane. Both mitochondria and sorting endosomes undergo major rearrangements leading to fission where they contact the ER. Sites of close apposition can also form between most of these organelles most pairwise combinations. First mentions of these contact sites can be found in papers published in the late 1950s mainly visualized using electron microscopy (EM) techniques. Copeland and Dalton described them as “highly specialized tubular form of endoplasmic reticulum in association with the mitochondria and apparently in turn, with the vascular border of the cell”.
LiMETER stands for light-inducible membrane-tethered peripheral endoplasmic reticulum (ER). LiMETER is an optogenetics tool designed to reversibly label cortical ER or the apposition between plasma membrane (PM) and endoplasmic reticulum (ER) membranes.
The ryanodine-inositol 1,4,5-triphosphate receptor Ca2+ channel (RIR-CaC) family includes Ryanodine receptors and Inositol trisphosphate receptors. Members of this family are large proteins, some exceeding 5000 amino acyl residues in length. This family belongs to the Voltage-gated ion channel (VIC) superfamily. Ry receptors occur primarily in muscle cell sarcoplasmic reticular (SR) membranes, and IP3 receptors occur primarily in brain cell endoplasmic reticular (ER) membranes where they effect release of Ca2+ into the cytoplasm upon activation (opening) of the channel. They are redox sensors, possibly providing a partial explanation for how they control cytoplasmic Ca2+. Ry receptors have been identified in heart mitochondria where they provide the main pathway for Ca2+ entry. Sun et al. (2011) have demonstrated oxygen-coupled redox regulation of the skeletal muscle ryanodine receptor-Ca2+ release channel (RyR1;TC# 1.A.3.1.2) by NADPH oxidase 4.
Anjana Rao is a cellular and molecular biologist of Indian ethnicity, working in the US. She uses immune cells as well as other types of cells to understand intracellular signaling and gene expression. Her research focuses on how signaling pathways control gene expression.
Patrick G. Hogan is a cellular and molecular biologist who studies how cellular signaling leads to gene expression. He obtained his bachelor’s degree from Harvard University and a PhD in neurobiology from Harvard Medical School. In 2010, he moved to the La Jolla Institute for Immunology in San Diego as a Professor in the Division of Signaling and Gene Expression. He is a Founder and Member of the Scientific Advisory Board, CalciMedica Inc, La Jolla, CA.














