
Phospholipids are a class of lipids whose molecule has a hydrophilic "head" containing a phosphate group and two hydrophobic "tails" derived from fatty acids, joined by an alcohol residue. Marine phospholipids typically have omega-3 fatty acids EPA and DHA integrated as part of the phospholipid molecule. The phosphate group can be modified with simple organic molecules such as choline, ethanolamine or serine.

Peripheral membrane proteins, or extrinsic membrane proteins, are membrane proteins that adhere only temporarily to the biological membrane with which they are associated. These proteins attach to integral membrane proteins, or penetrate the peripheral regions of the lipid bilayer. The regulatory protein subunits of many ion channels and transmembrane receptors, for example, may be defined as peripheral membrane proteins. In contrast to integral membrane proteins, peripheral membrane proteins tend to collect in the water-soluble component, or fraction, of all the proteins extracted during a protein purification procedure. Proteins with GPI anchors are an exception to this rule and can have purification properties similar to those of integral membrane proteins.

Phosphatidylinositol or inositol phospholipid is a biomolecule. It was initially called "inosite" when it was discovered by Léon Maquenne and Johann Joseph von Scherer in the late 19th century. It was discovered in bacteria but later also found in eukaryotes, and was found to be a signaling molecule.
Phosphatidic acids are anionic phospholipids important to cell signaling and direct activation of lipid-gated ion channels. Hydrolysis of phosphatidic acid gives rise to one molecule each of glycerol and phosphoric acid and two molecules of fatty acids. They constitute about 0.25% of phospholipids in the bilayer.

Phosphoinositide phospholipase C is a family of eukaryotic intracellular enzymes that play an important role in signal transduction processes. These enzymes belong to a larger superfamily of Phospholipase C. Other families of phospholipase C enzymes have been identified in bacteria and trypanosomes. Phospholipases C are phosphodiesterases.
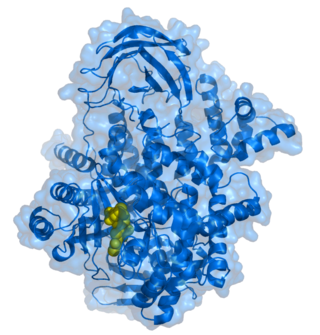
Phosphoinositide 3-kinases (PI3Ks), also called phosphatidylinositol 3-kinases, are a family of enzymes involved in cellular functions such as cell growth, proliferation, differentiation, motility, survival and intracellular trafficking, which in turn are involved in cancer.
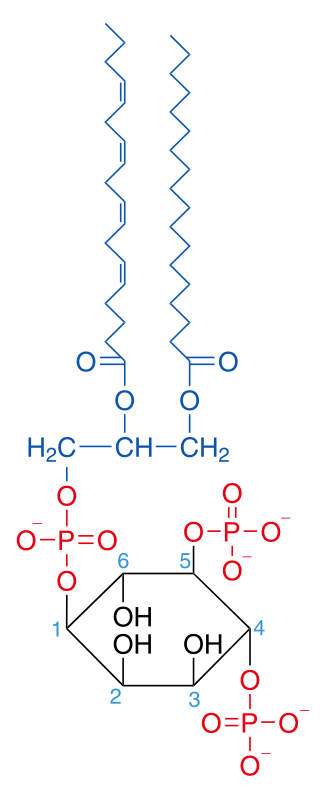
Phosphatidylinositol 4,5-bisphosphate or PtdIns(4,5)P2, also known simply as PIP2 or PI(4,5)P2, is a minor phospholipid component of cell membranes. PtdIns(4,5)P2 is enriched at the plasma membrane where it is a substrate for a number of important signaling proteins. PIP2 also forms lipid clusters that sort proteins.

Phosphatidylinositol 3-phosphate (PI3P) is a phospholipid found in cell membranes that helps to recruit a range of proteins, many of which are involved in protein trafficking, to the membranes. It is the product of both the class II and III phosphoinositide 3-kinases activity on phosphatidylinositol.
Phospholipase D (EC 3.1.4.4, lipophosphodiesterase II, lecithinase D, choline phosphatase, PLD; systematic name phosphatidylcholine phosphatidohydrolase) is an enzyme of the phospholipase superfamily that catalyses the following reaction
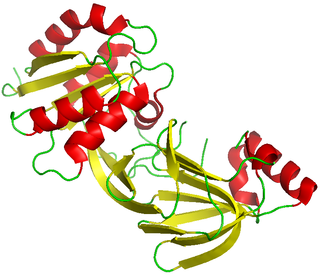
Diacylglycerol kinase is a family of enzymes that catalyzes the conversion of diacylglycerol (DAG) to phosphatidic acid (PA), utilizing ATP as a source of the phosphate. In non-stimulated cells, DGK activity is low, allowing DAG to be used for glycerophospholipid biosynthesis, but on receptor activation of the phosphoinositide pathway, DGK activity increases, driving the conversion of DAG to PA. As both lipids are thought to function as bioactive lipid signaling molecules with distinct cellular targets, DGK therefore occupies an important position, effectively serving as a switch by terminating the signalling of one lipid while simultaneously activating signalling by another.
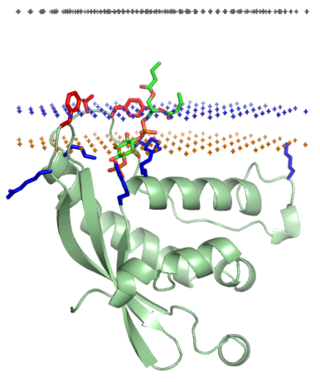
The PX domain is a phosphoinositide-binding structural domain involved in targeting of proteins to cell membranes.
Phosphatidylinositol transfer protein (PITP) or priming in exocytosis protein 3 (PEP3) is a ubiquitous cytosolic domain involved in transport of phospholipids from their site of synthesis in the endoplasmic reticulum and Golgi to other cell membranes.

The enzyme phosphatidate phosphatase (PAP, EC 3.1.3.4) is a key regulatory enzyme in lipid metabolism, catalyzing the conversion of phosphatidate to diacylglycerol:

Phospholipase C (PLC) is a class of membrane-associated enzymes that cleave phospholipids just before the phosphate group (see figure). It is most commonly taken to be synonymous with the human forms of this enzyme, which play an important role in eukaryotic cell physiology, in particular signal transduction pathways. Phospholipase C's role in signal transduction is its cleavage of phosphatidylinositol 4,5-bisphosphate (PIP2) into diacyl glycerol (DAG) and inositol 1,4,5-trisphosphate (IP3), which serve as second messengers. Activators of each PLC vary, but typically include heterotrimeric G protein subunits, protein tyrosine kinases, small G proteins, Ca2+, and phospholipids.

Phosphatidylinositol 4-kinase beta is an enzyme that in humans is encoded by the PI4KB gene.

Collagen type IV alpha-3-binding protein, also known as ceramide transfer protein (CERT) or StAR-related lipid transfer protein 11 (STARD11) is a protein that in humans is encoded by the COL4A3BP gene. The protein contains a pleckstrin homology domain at its amino terminus and a START domain towards the end of the molecule. It is a member of the StarD2 subfamily of START domain proteins.
Membrane contact sites (MCS) are close appositions between two organelles. Ultrastructural studies typically reveal an intermembrane distance in the order of the size of a single protein, as small as 10 nm or wider, with no clear upper limit. These zones of apposition are highly conserved in evolution. These sites are thought to be important to facilitate signalling, and they promote the passage of small molecules, including ions, lipids and reactive oxygen species. MCS are important in the function of the endoplasmic reticulum (ER), since this is the major site of lipid synthesis within cells. The ER makes close contact with many organelles, including mitochondria, Golgi, endosomes, lysosomes, peroxisomes, chloroplasts and the plasma membrane. Both mitochondria and sorting endosomes undergo major rearrangements leading to fission where they contact the ER. Sites of close apposition can also form between most of these organelles most pairwise combinations. First mentions of these contact sites can be found in papers published in the late 1950s mainly visualized using electron microscopy (EM) techniques. Copeland and Dalton described them as “highly specialized tubular form of endoplasmic reticulum in association with the mitochondria and apparently in turn, with the vascular border of the cell”.

Sec14 is a cytosolic protein found in yeast which plays a role in the regulation of several cellular functions, specifically those related to intracellular transport. Encoded by the Sec14 gene, Sec14p may transport phosphatidylinositol and phosphatidylcholine produced in the endoplasmic reticulum and the Golgi body to other cellular membranes. Additionally, Sec14p potentially plays a role in the localization of lipid raft proteins. Sec14p is an essential gene in yeast, and is homologous in function to phosphatidylinositol transfer protein in mammals. A conditional mutant with non-functional Sec14p presents with Berkeley bodies and deficiencies in protein secretion.
A FFAT motif is a protein sequence motif of six defined amino acids plus neighbouring residues that binds to proteins in the VAP protein family.
VAP proteins are conserved integral membrane proteins of the endoplasmic reticulum found in all eukaryotic cells. VAP stands for VAMP-associated protein, where VAMP stands for vesicle-associated membrane protein. Humans have two VAPs that consist of the essential Major Sperm Protein domain and linker plus transmembrane helix to attach to the ER: VAPA and VAPB. A third VAP-like protein is Motile sperm domain containing 2 (MOSPD2), which has all the elements of VAP, and like them binds FFAT motifs, but has at its N-terminus a CRAL-TRIO domain that can bind and transfer lipids.
















