
Histone methyltransferases (HMT) are histone-modifying enzymes, that catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues of histone proteins. The attachment of methyl groups occurs predominantly at specific lysine or arginine residues on histones H3 and H4. Two major types of histone methyltranferases exist, lysine-specific and arginine-specific. In both types of histone methyltransferases, S-Adenosyl methionine (SAM) serves as a cofactor and methyl donor group.
The genomic DNA of eukaryotes associates with histones to form chromatin. The level of chromatin compaction depends heavily on histone methylation and other post-translational modifications of histones. Histone methylation is a principal epigenetic modification of chromatin that determines gene expression, genomic stability, stem cell maturation, cell lineage development, genetic imprinting, DNA methylation, and cell mitosis.
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decrease transcription of genes, depending on which amino acids in the histones are methylated, and how many methyl groups are attached. Methylation events that weaken chemical attractions between histone tails and DNA increase transcription because they enable the DNA to uncoil from nucleosomes so that transcription factor proteins and RNA polymerase can access the DNA. This process is critical for the regulation of gene expression that allows different cells to express different genes.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.

DNA-directed RNA polymerases I, II, and III subunit RPABC1 is a protein that in humans is encoded by the POLR2E gene.

DNA-directed RNA polymerase II subunit RPB2 is an enzyme that in humans is encoded by the POLR2B gene.
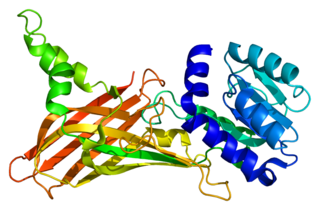
Protein arginine N-methyltransferase 1 is an enzyme that in humans is encoded by the PRMT1 gene. The HRMT1L2 gene encodes a protein arginine methyltransferase that functions as a histone methyltransferase specific for histone H4.

DNA (cytosine-5)-methyltransferase 3A (DNMT3A) is an enzyme that catalyzes the transfer of methyl groups to specific CpG structures in DNA, a process called DNA methylation. The enzyme is encoded in humans by the DNMT3A gene.

Glycylpeptide N-tetradecanoyltransferase 1 also known as myristoyl-CoA:protein N-myristoyltransferase 1 (NMT-1) is an enzyme that in humans is encoded by the NMT1 gene. It belongs to the protein N-terminal methyltransferase and glycylpeptide N-tetradecanoyltransferase family of enzymes.

Serine/threonine-protein kinase D3 (PKD3) or PKC-nu is an enzyme that in humans is encoded by the PRKD3 gene.

Heterogeneous nuclear ribonucleoprotein U-like protein 1 is a protein that in humans is encoded by the HNRNPUL1 gene.

Tubulin alpha-8 chain is a protein that in humans is encoded by the TUBA8 gene.
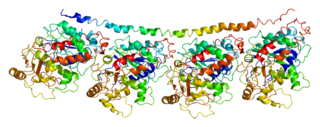
Tubulin alpha-1C chain is a protein that in humans is encoded by the TUBA1C gene.
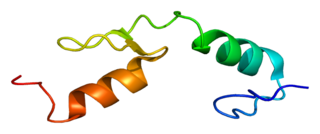
Zinc finger protein 40 is a protein that in humans is encoded by the HIVEP1 gene.
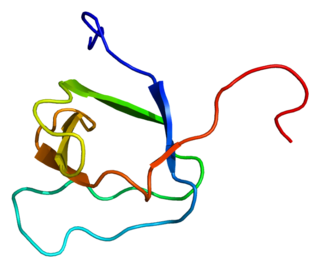
Protein arginine N-methyltransferase 2 is an enzyme that in humans is encoded by the PRMT2 gene.

TUBB1 is a gene that codes for the protein Tubulin beta-1 chain in humans.

tRNA (cytosine-5-)-methyltransferase is an enzyme that in humans is encoded by the TRDMT1 gene.
Protein arginine N-methyltransferase 3 is an enzyme that in humans is encoded by the PRMT3 gene.

HBx is a hepatitis B viral protein. It is 154 amino acids long and interferes with transcription, signal transduction, cell cycle progress, protein degradation, apoptosis and chromosomal stability in the host. It forms a heterodimeric complex with its cellular target protein, and this interaction dysregulates centrosome dynamics and mitotic spindle formation. It interacts with DDB1 redirecting the ubiquitin ligase activity of the CUL4-DDB1 E3 complexes, which are intimately involved in the intracellular regulation of DNA replication and repair, transcription and signal transduction.

For molecular biology in mammals, DNA demethylation causes replacement of 5-methylcytosine (5mC) in a DNA sequence by cytosine (C). DNA demethylation can occur by an active process at the site of a 5mC in a DNA sequence or, in replicating cells, by preventing addition of methyl groups to DNA so that the replicated DNA will largely have cytosine in the DNA sequence.

Euchromatic histone-lysine N-methyltransferase 1, also known as G9a-like protein (GLP), is a protein that in humans is encoded by the EHMT1 gene.





















