Related Research Articles
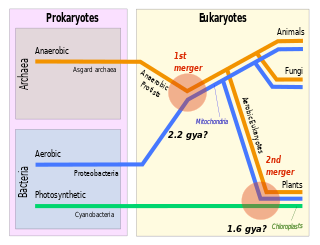
Symbiogenesis is the leading evolutionary theory of the origin of eukaryotic cells from prokaryotic organisms. The theory holds that mitochondria, plastids such as chloroplasts, and possibly other organelles of eukaryotic cells are descended from formerly free-living prokaryotes taken one inside the other in endosymbiosis. Mitochondria appear to be phylogenetically related to Rickettsiales bacteria, while chloroplasts are thought to be related to cyanobacteria.

RNA editing is a molecular process through which some cells can make discrete changes to specific nucleotide sequences within an RNA molecule after it has been generated by RNA polymerase. It occurs in all living organisms and is one of the most evolutionarily conserved properties of RNAs. RNA editing may include the insertion, deletion, and base substitution of nucleotides within the RNA molecule. RNA editing is relatively rare, with common forms of RNA processing not usually considered as editing. It can affect the activity, localization as well as stability of RNAs, and has been linked with human diseases.

F-box proteins are proteins containing at least one F-box domain. The first identified F-box protein is one of three components of the SCF complex, which mediates ubiquitination of proteins targeted for degradation by the 26S proteasome.
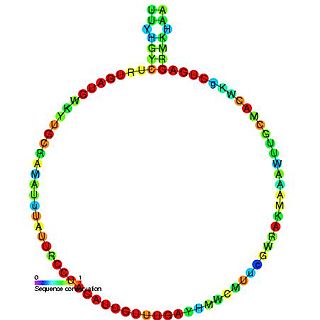
In molecular biology, snoRNA U34 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

CUG triplet repeat, RNA binding protein 1, also known as CUGBP1, is a protein which in humans is encoded by the CUGBP1 gene.

CUGBP, Elav-like family member 2, also known as Etr-3 is a protein that in humans is encoded by the CELF2 gene.

Transcription elongation regulator 1, also known as TCERG1, is a protein which in humans is encoded by the TCERG1 gene.

40S ribosomal protein S16' is a protein that in humans is encoded by the RPS16 gene.

60S ribosomal protein L14 is a protein that in humans is encoded by the RPL14 gene.

60S ribosomal protein L12 is a protein that in humans is encoded by the RPL12 gene.

60S ribosomal protein L28 is a protein that in humans is encoded by the RPL28 gene.

60S ribosomal protein L13a is a protein that in humans is encoded by the RPL13A gene.

Leucine-rich PPR motif-containing protein, mitochondrial is a protein that in humans is encoded by the LRPPRC gene. Transcripts ranging in size from 4.8 to 7.0 kb which result from alternative polyadenylation have been reported for this gene.

40S ribosomal protein S8 is a protein that in humans is encoded by the RPS8 gene.

Pleiotropic regulator 1 is a protein that in humans is encoded by the PLRG1 gene.

60S ribosomal protein L37 is a protein that in humans is encoded by the RPL37 gene.
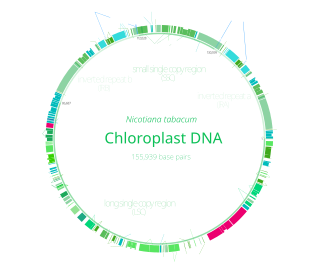
Chloroplast DNA (cpDNA) is the DNA located in chloroplasts, which are photosynthetic organelles located within the cells of some eukaryotic organisms. Chloroplasts, like other types of plastid, contain a genome separate from that in the cell nucleus. The existence of chloroplast DNA was identified biochemically in 1959, and confirmed by electron microscopy in 1962. The discoveries that the chloroplast contains ribosomes and performs protein synthesis revealed that the chloroplast is genetically semi-autonomous. The first complete chloroplast genome sequences were published in 1986, Nicotiana tabacum (tobacco) by Sugiura and colleagues and Marchantia polymorpha (liverwort) by Ozeki et al. Since then, a great number of chloroplast DNAs from various species have been sequenced.
Alice Barkan is an American molecular biologist and a professor of biology at the University of Oregon. She is known for her work on chloroplast gene regulation and protein synthesis.
Jian-Kang Zhu is a plant scientist, researcher and academic. He is a Senior Principal Investigator in the Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences (CAS). He is also the Academic Director of CAS Center of Excellence in Plant Sciences.

Maureen Hanson is an American molecular biologist and Liberty Hyde Bailey Professor in the Department of Molecular Biology and Genetics at Cornell University in Ithaca, New York. She is a joint member of the Section of Plant Biology and Director of the Center for Enervating Neuroimmune Disease. Her research concerns gene expression in chloroplasts and mitochondria, photosynthesis, and the molecular basis of the disease Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS).
References
- ↑ Mingler MK, Hingst AM, Clement SL, Yu LE, Reifur L, Koslowsky DJ (November 2006). "Identification of pentatricopeptide repeat proteins in Trypanosoma brucei". Mol. Biochem. Parasitol. 150 (1): 37–45. doi:10.1016/j.molbiopara.2006.06.006. PMID 16837079.
- 1 2 3 Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, Lecharny A, Le Ret M, Martin-Magniette ML, Mireau H, Peeters N, Renou JP, Szurek B, Taconnat L, Small I (August 2004). "Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis". Plant Cell. 16 (8): 2089–103. doi:10.1105/tpc.104.022236. PMC 519200 . PMID 15269332.
- ↑ O'Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I (June 2008). "On the expansion of the pentatricopeptide repeat gene family in plants". Mol. Biol. Evol. 25 (6): 1120–8. doi: 10.1093/molbev/msn057 . PMID 18343892.
- ↑ Kotera E, Tasaka M, Shikanai T (January 2005). "A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts". Nature. 433 (7023): 326–30. Bibcode:2005Natur.433..326K. doi:10.1038/nature03229. PMID 15662426. S2CID 4416316.
- ↑ Takenaka M, Verbitskiy D, Zehrmann A, Brennicke A (June 2010). "Reverse genetic screening identifies five E-class PPR-proteins involved in RNA editing in mitochondria of Arabidopsis Thaliana". J Biol Chem. 285 (35): 27122–27129. doi: 10.1074/jbc.M110.128611 . PMC 2930711 . PMID 20566637.
- ↑ Yin P, Li Q, Yan C, Liu Y, Liu J, Yu F, et al. (December 2013). "Structural basis for the modular recognition of single-stranded RNA by PPR proteins". Nature. 504 (7478): 168–71. Bibcode:2013Natur.504..168Y. doi:10.1038/nature12651. PMID 24162847. S2CID 4471801.