In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.
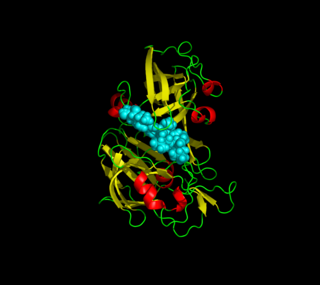
Plasmepsins are a class of at least 10 enzymes produced by the Plasmodium falciparum parasite. There are ten different isoforms of these proteins and ten genes coding them respectively in Plasmodium. It has been suggested that the plasmepsin family is smaller in other human Plasmodium species. Expression of Plm I, II, IV, V, IX, X and HAP occurs in the erythrocytic cycle, and expression of Plm VI, VII, VIII, occurs in the exoerythrocytic cycle. Through their haemoglobin-degrading activity, they are an important cause of symptoms in malaria sufferers. Consequently, this family of enzymes is a potential target for antimalarial drugs.

β-Glucocerebrosidase is an enzyme with glucosylceramidase activity that is needed to cleave, by hydrolysis, the beta-glucosidic linkage of the chemical glucocerebroside, an intermediate in glycolipid metabolism that is abundant in cell membranes. It is localized in the lysosome, where it remains associated with the lysosomal membrane. β-Glucocerebrosidase is 497 amino acids in length and has a molecular weight of 59,700 Daltons.
Aspergillopepsin I is an enzyme. This enzyme catalyses the following chemical reaction
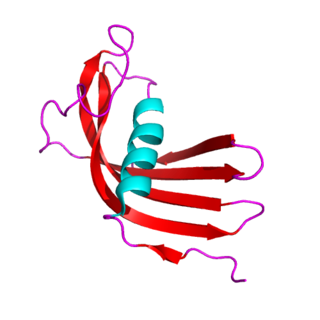
The cystatins are a family of cysteine protease inhibitors which share a sequence homology and a common tertiary structure of an alpha helix lying on top of an anti-parallel beta sheet. The family is subdivided as described below.

Aspartic proteases are a catalytic type of protease enzymes that use an activated water molecule bound to one or more aspartate residues for catalysis of their peptide substrates. In general, they have two highly conserved aspartates in the active site and are optimally active at acidic pH. Nearly all known aspartyl proteases are inhibited by pepstatin.

Thermolysin is a thermostable neutral metalloproteinase enzyme produced by the Gram-positive bacteria Bacillus thermoproteolyticus. It requires one zinc ion for enzyme activity and four calcium ions for structural stability. Thermolysin specifically catalyzes the hydrolysis of peptide bonds containing hydrophobic amino acids. However thermolysin is also widely used for peptide bond formation through the reverse reaction of hydrolysis. Thermolysin is the most stable member of a family of metalloproteinases produced by various Bacillus species. These enzymes are also termed 'neutral' proteinases or thermolysin -like proteinases (TLPs).

Prosaposin, also known as PSAP, is a protein which in humans is encoded by the PSAP gene.

In molecular biology Proteinase K is a broad-spectrum serine protease. The enzyme was discovered in 1974 in extracts of the fungus Engyodontium album. Proteinase K is able to digest hair (keratin), hence, the name "Proteinase K". The predominant site of cleavage is the peptide bond adjacent to the carboxyl group of aliphatic and aromatic amino acids with blocked alpha amino groups. It is commonly used for its broad specificity. This enzyme belongs to Peptidase family S8 (subtilisin). The molecular weight of Proteinase K is 28,900 daltons.
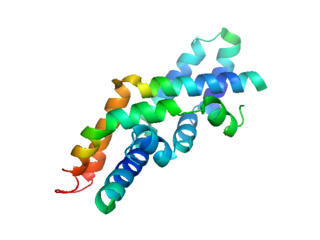
The saposin domains refers to two evolutionally-conserved protein domains found in saposin and related proteins (SAPLIP). Saposins are small lysosomal proteins that serve as activators of various lysosomal lipid-degrading enzymes. They probably act by isolating the lipid substrate from the membrane surroundings, thus making it more accessible to the soluble degradative enzymes. All mammalian saposins are synthesized as a single precursor molecule (prosaposin) which contains four Saposin-B domains, yielding the active saposins after proteolytic cleavage, and two Saposin-A domains that are removed in the activation reaction.

Cathepsin E is an enzyme that in humans is encoded by the CTSE gene. The enzyme is also known as slow-moving proteinase, erythrocyte membrane aspartic proteinase, SMP, EMAP, non-pepsin proteinase, cathepsin D-like acid proteinase, cathepsin E-like acid proteinase, cathepsin D-type proteinase) is an enzyme.
Nepenthesin is an aspartic protease of plant origin that has so far been identified in the pitcher secretions of Nepenthes and in the leaves of Drosera peltata. It is similar to pepsin, but differs in that it also cleaves on either side of Asp residues and at Lys┼Arg. While more pH and temperature stable than porcine pepsin A, it is considerably less stable in urea or guanidine hydrochloride. It is the only known protein with such a stability profile.
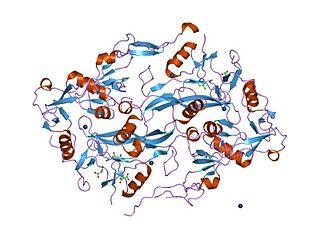
The Kazal domain is an evolutionary conserved protein domain usually indicative of serine protease inhibitors. However, kazal-like domains are also seen in the extracellular part of agrins, which are not known to be protease inhibitors.

In molecular biology, the CDC48 N-terminal domain is a protein domain found in AAA ATPases including cell division protein 48 (CDC48), VCP-like ATPase and N-ethylmaleimide sensitive fusion protein. It is a substrate recognition domain which binds polypeptides, prevents protein aggregation, and catalyses refolding of permissive substrates. It is composed of two equally sized subdomains. The amino-terminal subdomain (CDC48_N) forms a double-psi beta-barrel whose pseudo-twofold symmetry is mirrored by an internal sequence repeat of 42 residues. The carboxy-terminal subdomain (CDC48_2) forms a novel six-stranded beta-clam fold. Together these subdomains form a kidney-shaped structure, in close agreement with results from electron microscopy. CDC48_N is related to numerous proteins including prokaryotic transcription factors, metabolic enzymes, the protease cofactors UFD1 and PrlF, and aspartic proteinases.
Streptogrisin B is an enzyme. This enzyme catalyses the following chemical reaction

Aspergilloglutamic peptidase, also called aspergillopepsin II is a proteolytic enzyme. The enzyme was previously thought be an aspartic protease, but it was later shown to be a glutamic protease with a catalytic Glu residue at the active site, and was therefore renamed aspergilloglutamic peptidase.
Rhizopuspepsin is an enzyme. This enzyme catalyses the following chemical reaction
Mucorpepsin is an enzyme. This enzyme catalyses the following chemical reaction
Rhodotorulapepsin is an enzyme. This enzyme catalyses the following chemical reaction

Scytalidocarboxyl peptidase B, also known as Scytalidoglutamic peptidase and Scytalidopepsin B is a proteolytic enzyme. It was previously thought to be an aspartic protease, but determination of its molecular structure showed it to belong a novel group of proteases, glutamic protease.














