Metachromatic leukodystrophy (MLD) is a lysosomal storage disease which is commonly listed in the family of leukodystrophies as well as among the sphingolipidoses as it affects the metabolism of sphingolipids. Leukodystrophies affect the growth and/or development of myelin, the fatty covering which acts as an insulator around nerve fibers throughout the central and peripheral nervous systems. MLD involves cerebroside sulfate accumulation. Metachromatic leukodystrophy, like most enzyme deficiencies, has an autosomal recessive inheritance pattern.

GM2-gangliosidosis, AB variant is a rare, autosomal recessive metabolic disorder that causes progressive destruction of nerve cells in the brain and spinal cord. It has a similar pathology to Sandhoff disease and Tay–Sachs disease. The three diseases are classified together as the GM2 gangliosidoses, because each disease represents a distinct molecular point of failure in the activation of the same enzyme, beta-hexosaminidase. AB variant is caused by a failure in the gene that makes an enzyme cofactor for beta-hexosaminidase, called the GM2 activator.

Angiogenin (ANG) also known as ribonuclease 5 is a small 123 amino acid protein that in humans is encoded by the ANG gene. Angiogenin is a potent stimulator of new blood vessels through the process of angiogenesis. Ang hydrolyzes cellular RNA, resulting in modulated levels of protein synthesis and interacts with DNA causing a promoter-like increase in the expression of rRNA. Ang is associated with cancer and neurological disease through angiogenesis and through activating gene expression that suppresses apoptosis.

Hexosaminidase is an enzyme involved in the hydrolysis of terminal N-acetyl-D-hexosamine residues in N-acetyl-β-D-hexosaminides.

Arylsulfatase A is an enzyme that breaks down sulfatides, namely cerebroside 3-sulfate into cerebroside and sulfate. In humans, arylsulfatase A is encoded by the ARSA gene.

Tissue alpha-L-fucosidase is an enzyme that in humans is encoded by the FUCA1 gene.

ADP-ribosylation factor 1 is a protein that in humans is encoded by the ARF1 gene.

GM2 ganglioside activator also known as GM2A is a protein which in humans is encoded by the GM2A gene.

Sphingosine-1-phosphate receptor 2, also known as S1PR2 or S1P2, is a human gene which encodes a G protein-coupled receptor which binds the lipid signaling molecule sphingosine 1-phosphate (S1P).

Probable G-protein coupled receptor 135 is a protein that in humans is encoded by the GPR135 gene.
In enzymology, a dephospho-[reductase kinase] kinase is an enzyme that catalyzes the chemical reaction
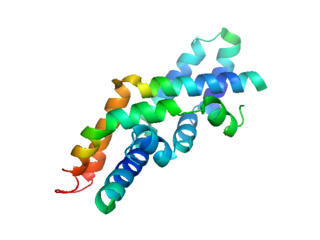
The saposin domains refers to two evolutionally-conserved protein domains found in saposin and related proteins (SAPLIP). Saposins are small lysosomal proteins that serve as activators of various lysosomal lipid-degrading enzymes. They probably act by isolating the lipid substrate from the membrane surroundings, thus making it more accessible to the soluble degradative enzymes. All mammalian saposins are synthesized as a single precursor molecule (prosaposin) which contains four Saposin-B domains, yielding the active saposins after proteolytic cleavage, and two Saposin-A domains that are removed in the activation reaction.

5'-AMP-activated protein kinase catalytic subunit alpha-2 is an enzyme that in humans is encoded by the PRKAA2 gene.

The human gene UBR1 encodes the enzyme ubiquitin-protein ligase E3 component n-recognin 1.
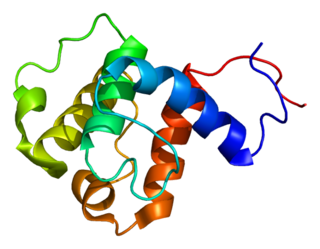
NEDD9-interacting protein with calponin homology and LIM domains is a protein that in humans is encoded by the MICAL1 gene.

Protein CLN8 is a protein that in humans is encoded by the CLN8 gene.

40S ribosomal protein S18 is a protein that in humans is encoded by the RPS18 gene.

Tensin-like C1 domain-containing phosphatase is an enzyme that in humans is encoded by the TENC1 gene.
Richard I. Morimoto is a Japanese American molecular biologist. He is the Bill and Gayle Cook Professor of Biology and Director of the Rice Institute for Biomedical Research at Northwestern University.
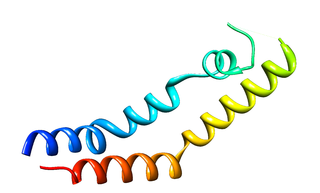
The plant-specific insert (PSI) or plant-specific sequence (PSS) is an independent domain, exclusively found in plants, consisting of approximately 100 residues, found on the C-terminal lobe on some aspartic proteases (AP) called phytepsins. The PSI, as an independent entity separate from its parent AP, is homologous to saposin and belongs to the saposin-like protein family (SAPLIP).































