
A lysosome is a membrane-bound organelle found in many animal cells. They are spherical vesicles that contain hydrolytic enzymes that can break down many kinds of biomolecules. A lysosome has a specific composition, of both its membrane proteins, and its lumenal proteins. The lumen's pH (~4.5–5.0) is optimal for the enzymes involved in hydrolysis, analogous to the activity of the stomach. Besides degradation of polymers, the lysosome is involved in various cell processes, including secretion, plasma membrane repair, apoptosis, cell signaling, and energy metabolism.

Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion.

SNARE proteins – "SNAPREceptor" – are a large protein family consisting of at least 24 members in yeasts and more than 60 members in mammalian cells. The primary role of SNARE proteins is to mediate vesicle fusion – the fusion of vesicles with the target membrane; this notably mediates exocytosis, but can also mediate the fusion of vesicles with membrane-bound compartments. The best studied SNAREs are those that mediate the neurotransmitter release of synaptic vesicles in neurons. These neuronal SNAREs are the targets of the neurotoxins responsible for botulism and tetanus produced by certain bacteria.
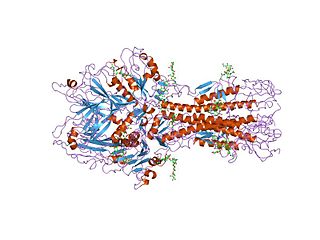
Hemagglutinin esterase (HEs) is a glycoprotein that certain enveloped viruses possess and use as invading mechanism. HEs helps in the attachment and destruction of certain sialic acid receptors that are found on the host cell surface. Viruses that possess HEs include Influenza C virus, toroviruses, and coronaviruses. HEs is a dimer transmembrane protein consisting of two monomers, each monomer is made of three domains. The three domains are: membrane fusion, esterase, and receptor binding domains.

Spectrin is a cytoskeletal protein that lines the intracellular side of the plasma membrane in eukaryotic cells. Spectrin forms pentagonal or hexagonal arrangements, forming a scaffold and playing an important role in maintenance of plasma membrane integrity and cytoskeletal structure. The hexagonal arrangements are formed by tetramers of spectrin subunits associating with short actin filaments at either end of the tetramer. These short actin filaments act as junctional complexes allowing the formation of the hexagonal mesh. The protein is named spectrin since it was first isolated as a major protein component of human red blood cells which had been treated with mild detergents; the detergents lysed the cells and the hemoglobin and other cytoplasmic components were washed out. In the light microscope the basic shape of the red blood cell could still be seen as the spectrin-containing submembranous cytoskeleton preserved the shape of the cell in outline. This became known as a red blood cell "ghost" (spectre), and so the major protein of the ghost was named spectrin.

β-Glucocerebrosidase is an enzyme with glucosylceramidase activity that is needed to cleave, by hydrolysis, the beta-glucosidic linkage of the chemical glucocerebroside, an intermediate in glycolipid metabolism that is abundant in cell membranes. It is localized in the lysosome, where it remains associated with the lysosomal membrane. β-Glucocerebrosidase is 497 amino acids in length and has a molecular weight of 59,700 Daltons.

Sterol regulatory element-binding proteins (SREBPs) are transcription factors that bind to the sterol regulatory element DNA sequence TCACNCCAC. Mammalian SREBPs are encoded by the genes SREBF1 and SREBF2. SREBPs belong to the basic-helix-loop-helix leucine zipper class of transcription factors. Unactivated SREBPs are attached to the nuclear envelope and endoplasmic reticulum membranes. In cells with low levels of sterols, SREBPs are cleaved to a water-soluble N-terminal domain that is translocated to the nucleus. These activated SREBPs then bind to specific sterol regulatory element DNA sequences, thus upregulating the synthesis of enzymes involved in sterol biosynthesis. Sterols in turn inhibit the cleavage of SREBPs and therefore synthesis of additional sterols is reduced through a negative feed back loop.
Phospholipase D (PLD) is an enzyme of the phospholipase superfamily. Phospholipases occur widely, and can be found in a wide range of organisms, including bacteria, yeast, plants, animals, and viruses. Phospholipase D's principal substrate is phosphatidylcholine, which it hydrolyzes to produce the signal molecule phosphatidic acid (PA), and soluble choline. Plants contain numerous genes that encode various PLD isoenzymes, with molecular weights ranging from 90 to 125 kDa. Mammalian cells encode two isoforms of phospholipase D: PLD1 and PLD2. Phospholipase D is an important player in many physiological processes, including membrane trafficking, cytoskeletal reorganization, receptor-mediated endocytosis, exocytosis, and cell migration. Through these processes, it has been further implicated in the pathophysiology of multiple diseases: in particular the progression of Parkinson's and Alzheimer's, as well as various cancers. PLD may also help set the threshold for sensitivity to anesthetia and mechanical force.

Gamma secretase is a multi-subunit protease complex, itself an integral membrane protein, that cleaves single-pass transmembrane proteins at residues within the transmembrane domain. Proteases of this type are known as intramembrane proteases. The most well-known substrate of gamma secretase is amyloid precursor protein, a large integral membrane protein that, when cleaved by both gamma and beta secretase, produces a short 37-43 amino acid peptide called amyloid beta whose abnormally folded fibrillar form is the primary component of amyloid plaques found in the brains of Alzheimer's disease patients. Gamma secretase is also critical in the related processing of several other type I integral membrane proteins, such as Notch, ErbB4, E-cadherin, N-cadherin, ephrin-B2, or CD44.

Sterol regulatory element-binding protein cleavage-activating protein, also known as SREBP cleavage-activating protein or SCAP is a protein that in humans is encoded by the SCAP gene.

Surfactant protein B is an essential lipid-associated protein found in pulmonary surfactant. Without it, the lung would not be able to inflate after a deep breath out. It rearranges lipid molecules in the fluid lining the lung so that tiny air sacs in the lung, called alveoli, can more easily inflate.

Prosaposin, also known as PSAP, is a protein which in humans is encoded by the PSAP gene.

Proteinase-activated receptor 1 (PAR1) also known as Protease-activated receptor 1 or coagulation factor II (thrombin) receptor is a protein that in humans is encoded by the F2R gene. PAR1 is a G protein-coupled receptor and one of four protease-activated receptors involved in the regulation of thrombotic response. Highly expressed in platelets and endothelial cells, PAR1 plays a key role in mediating the interplay between coagulation and inflammation, which is important in the pathogenesis of inflammatory and fibrotic lung diseases. It is also involved both in disruption and maintenance of endothelial barrier integrity, through interaction with either thrombin or activated protein C, respectively.

In enzymology, an acyloxyacyl hydrolase is an enzyme that catalyzes the chemical reaction

Phospholipase C (PLC) is a class of membrane-associated enzymes that cleave phospholipids just before the phosphate group (see figure). It is most commonly taken to be synonymous with the human forms of this enzyme, which play an important role in eukaryotic cell physiology, in particular signal transduction pathways. There are thirteen kinds of mammalian phospholipase C that are classified into six isotypes (β, γ, δ, ε, ζ, η) according to structure. Each PLC has unique and overlapping controls over expression and subcellular distribution. Activators of each PLC vary, but typically include heterotrimeric G protein subunits, protein tyrosine kinases, small G proteins, Ca2+, and phospholipids.
Angiogenesis is the process of forming new blood vessels from existing blood vessels. It is a highly complex process involving extensive interplay between cells, soluble factors, and the extracellular matrix (ECM). Angiogenesis is critical during normal physiological development, but it also occurs in adults during inflammation, wound healing, ischemia, and in pathological conditions such as rheumatoid arthritis, hemangioma, and tumor growth. Proteolysis has been indicated as one of the first and most sustained activities involved in the formation of new blood vessels. Numerous proteases including matrix metalloproteases (MMPs), a disintegrin and metalloprotease domain (ADAM), a disintegrin and metalloprotease domain with throbospondin motifs (ADAMTS), and cysteine and serine proteases are involved in angiogenesis. This article focuses on the important and diverse roles that these proteases play in the regulation of angiogenesis.
Surfactant metabolism dysfunction is a condition where pulmonary surfactant is insufficient for adequate respiration. Surface tension at the liquid-air interphase in the alveoli makes the air sacs prone to collapsing post expiration. This is due to the fact that water molecules in the liquid-air surface of alveoli are more attracted to one another than they are to molecules in the air. For sphere-like structures like alveoli, water molecules line the inner walls of the air sacs and stick tightly together through hydrogen bonds. These intermolecular forces put great restraint on the inner walls of the air sac, tighten the surface all together, and unyielding to stretch for inhalation. Thus, without something to alleviate this surface tension, alveoli can collapse and cannot be filled up again. Surfactant is essential mixture that is released into the air-facing surface of inner walls of air sacs to lessen the strength of surface tension. This mixture inserts itself among water molecules and breaks up hydrogen bonds that hold the tension. Multiple lung diseases, like ISD or RDS, in newborns and late-onsets cases have been linked to dysfunction of surfactant metabolism.
The plant-specific insert (PSI) or plant-specific sequence (PSS) is an independent domain, exclusively found in plants, consisting of approximately 100 residues, found on the C-terminal lobe on some aspartic proteases (AP) called phytepsins. The PSI, as an independent entity separate from its parent AP, is homologous to saposin and belongs to the saposin-like protein family (SAPLIP).

Chaperone-mediated autophagy (CMA) refers to the chaperone-dependent selection of soluble cytosolic proteins that are then targeted to lysosomes and directly translocated across the lysosome membrane for degradation. The unique features of this type of autophagy are the selectivity on the proteins that are degraded by this pathway and the direct shuttling of these proteins across the lysosomal membrane without the requirement for the formation of additional vesicles.

Gasdermin D (GSDMD) is a protein that in humans is encoded by the GSDMD gene on chromosome 8. It belongs to the gasdermin family which is conserved among all vertebrates and comprises six members, GSDMA, GSDMB, GSDMC, GSDMD, GSDME (DFNA5) and DFNB59 (Pejvakin). Members of the gasdermin family are mainly expressed in epithelial tissues and appear to play a role in regulation of epithelial proliferation and differentiation. GSDMA, GSDMC, GSDMD and DFNA5 have been suggested to act as tumour suppressors.


















