Related Research Articles

A flagellum is a hairlike appendage that protrudes from certain plant and animal sperm cells, and from a wide range of microorganisms to provide motility. Many protists with flagella are termed as flagellates.

The cytoskeleton is a complex, dynamic network of interlinking protein filaments present in the cytoplasm of all cells, including those of bacteria and archaea. In eukaryotes, it extends from the cell nucleus to the cell membrane and is composed of similar proteins in the various organisms. It is composed of three main components, microfilaments, intermediate filaments and microtubules, and these are all capable of rapid growth or disassembly dependent on the cell's requirements.

ATP synthase is a protein that catalyzes the formation of the energy storage molecule adenosine triphosphate (ATP) using adenosine diphosphate (ADP) and inorganic phosphate (Pi). It is classified under ligases as it changes ADP by the formation of P-O bond (phosphodiester bond). ATP synthase is a molecular machine. The overall reaction catalyzed by ATP synthase is:

Vibrio is a genus of Gram-negative bacteria, possessing a curved-rod (comma) shape, several species of which can cause foodborne infection, usually associated with eating undercooked seafood. Being highly salt tolerant and unable to survive in fresh water, Vibrio spp. are commonly found in various salt water environments. Vibrio spp. are facultative anaerobes that test positive for oxidase and do not form spores. All members of the genus are motile. They are able to have polar or lateral flagellum with or without sheaths. Vibrio species typically possess two chromosomes, which is unusual for bacteria. Each chromosome has a distinct and independent origin of replication, and are conserved together over time in the genus. Recent phylogenies have been constructed based on a suite of genes.

Dyneins are a family of cytoskeletal motor proteins that move along microtubules in cells. They convert the chemical energy stored in ATP to mechanical work. Dynein transports various cellular cargos, provides forces and displacements important in mitosis, and drives the beat of eukaryotic cilia and flagella. All of these functions rely on dynein's ability to move towards the minus-end of the microtubules, known as retrograde transport; thus, they are called "minus-end directed motors". In contrast, most kinesin motor proteins move toward the microtubules' plus-end, in what is called anterograde transport.
In cellular biology, membrane transport refers to the collection of mechanisms that regulate the passage of solutes such as ions and small molecules through biological membranes, which are lipid bilayers that contain proteins embedded in them. The regulation of passage through the membrane is due to selective membrane permeability - a characteristic of biological membranes which allows them to separate substances of distinct chemical nature. In other words, they can be permeable to certain substances but not to others.
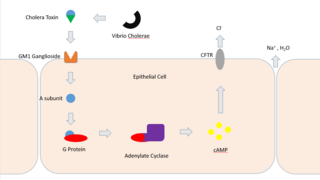
Cholera toxin is AB5 multimeric protein complex secreted by the bacterium Vibrio cholerae. CTX is responsible for the massive, watery diarrhea characteristic of cholera infection. It is a member of the Heat-labile enterotoxin family.
The sodium/phosphate cotransporter is a member of the phosphate:Na+ symporter (PNaS) family within the TOG Superfamily of transport proteins as specified in the Transporter Classification Database (TCDB).

Kinesin-like protein KIF3B is a protein that in humans is encoded by the KIF3B gene. KIF3B is an N-type protein that complexes with two other kinesin proteins to form two-headed anterograde motors. First, KIF3B forms a heterodimer with KIF3A ; (KIF3A/3B), that is membrane-bound and has ATPase activity. Then KIFAP3 binds to the tail domain to form a heterotrimeric motor. This motor has a plus end-directed microtubule sliding activity that exhibits a velocity of ∼0.3 μm/s a. There are 14 kinesin protein families in the kinesin superfamily and KIF3B is part of the Kinesin-2 family, of kinesins that can all form heterotrimeric complexes. Expression of the three motor subunits is ubiquitous. The KIG3A/3B/KAP3 motors can transport 90 to 160 nm in diameter organelles.

Multidrug and toxin extrusion protein 2 is a protein which in humans is encoded by the SLC47A2 gene.

Bacterial motility is the ability of bacteria to move independently using metabolic energy. Most motility mechanisms which evolved among bacteria also evolved in parallel among the archaea. Most rod-shaped bacteria can move using their own power, which allows colonization of new environments and discovery of new resources for survival. Bacterial movement depends not only on the characteristics of the medium, but also on the use of different appendages to propel. Swarming and swimming movements are both powered by rotating flagella. Whereas swarming is a multicellular 2D movement over a surface and requires the presence of surfactants, swimming is movement of individual cells in liquid environments.

Several organisms are capable of rolling locomotion. However, true wheels and propellers—despite their utility in human vehicles—do not seem to play a significant role in the movement of living things. Biologists have offered several explanations for the apparent absence of biological wheels, and wheeled creatures have appeared often in speculative fiction.
Multi-antimicrobial extrusion protein (MATE) also known as multidrug and toxin extrusion or multidrug and toxic compound extrusion is a family of proteins which function as drug/sodium or proton antiporters.
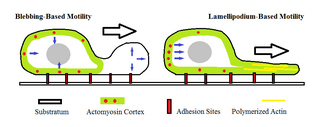
Amoeboid movement is the most typical mode of locomotion in adherent eukaryotic cells. It is a crawling-like type of movement accomplished by protrusion of cytoplasm of the cell involving the formation of pseudopodia ("false-feet") and posterior uropods. One or more pseudopodia may be produced at a time depending on the organism, but all amoeboid movement is characterized by the movement of organisms with an amorphous form that possess no set motility structures.
Motility protein A, MotA, is a bacterial protein that is encoded by the motA gene. It is a component of the flagellar motor. More specifically, MotA and MotB make the stator of a H+ driven bacterial flagella and surround the rotor as a ring of about 8–10 particles. MotA and MotB are integral membrane proteins. MotA has four transmembrane domains.
Motility protein B also known as MotB is a bacterial protein that is encoded by the motB gene. It's a component of the flagellar motor. More specifically, MotA and MotB makes the stator of a flagellum and surround the rotor as a ring of about 8-10 particles. MotA and MotB are integral membrane proteins. While both MotA and MotB surround the MS ring, MotB also anchors MotA to cell wall peptidoglycan. These two proteins form pores that harvest energy for flagellar mechanical movement by proton motive force (PMF) across the membrane. Cellular metabolic processes such as the electron transport chain move protons outside the cell, creating more protons and more positive charge in the extracellular space. When the protons flow back into the cell through MotA and MotB along concentration and charge gradients, they release energy that is used for flagellar rotation. The speed of the flagellar motor is dependent on the magnitude of the PMF acting on MotA and MotB.
PomA is a protein that is part of the stator in Na+ driven bacterial flagella. It has a high degree of homology to MotA, and Rhodobacter sphaeroides MotA can functionally complement a non-motile Vibrio alginolyticus with a non-functional pomA gene.

In molecular biology, the OmpA domain is a conserved protein domain with a beta/alpha/beta/alpha-beta(2) structure found in the C-terminal region of many Gram-negative bacterial outer membrane proteins, such as porin-like integral membrane proteins, small lipid-anchored proteins, and MotB proton channels. The N-terminal half of these proteins is variable although some of the proteins in this group have the OmpA-like transmembrane domain at the N terminus. OmpA from Escherichia coli is required for pathogenesis, and can interact with host receptor molecules. MotB serve two functions in E. coli, the MotA(4)-MotB(2) complex attaches to the cell wall via MotB to form the stator of the flagellar motor, and the MotA-MotB complex couples the flow of ions across the cell membrane to movement of the rotor.
The archaellum is a unique structure on the cell surface of many archaea, that allows for swimming motility. The archaellum consists of a rigid helical filament that is attached to the cell membrane by a molecular motor. This molecular motor – composed of cytosolic, membrane, and pseudo-periplasmic proteins – is responsible for the assembly of the filament and, once assembled, for its rotation. The rotation of the filament propels archaeal cells in liquid medium, in a manner similar to the propeller of a boat. The bacterial analog of the archaellum is the flagellum, which is also responsible for their swimming motility and can also be compared to a rotating corkscrew. Although the movement of archaella and flagella is sometimes described as "whip-like", this is incorrect, as only cilia from Eukaryotes move in this manner. Indeed, even "flagellum" is a misnomer, as bacterial flagella work are also propeller-like structures

Bacterial secretion systems are protein complexes present on the cell membranes of bacteria for secretion of substances. Specifically, they are the cellular devices used by pathogenic bacteria to secrete their virulence factors to invade the host cells. They can be classified into different types based on their specific structure, composition and activity. Generally, proteins can be secreted through two different processes. One process is a one-step mechanism in which proteins from the cytoplasm of bacteria are transported and delivered directly through the cell membrane into the host cell. Another involves a two-step activity in which the proteins are first transported out of the inner cell membrane, then deposited in the periplasm, and finally through the outer cell membrane into the host cell.
References
- ↑ Asai, Yukako; Kawagishi, Ikuro; Sockett, R. Elizabeth; Homma, Michio (October 1999). "Hybrid motor with H+- and Na+-driven components can rotate Vibrio polar flagella by using sodium ions". Journal of Bacteriology. 181 (20): 6332–38. doi:10.1128/JB.181.20.6332-6338.1999. PMC 103767 . PMID 10515922.