Related Research Articles

A mass spectrum is a type of plot of the ion signal as a function of the mass-to-charge ratio. These spectra are used to determine the elemental or isotopic signature of a sample, the masses of particles and of molecules, and to elucidate the chemical identity or structure of molecules and other chemical compounds.

Electrospray ionization (ESI) is a technique used in mass spectrometry to produce ions using an electrospray in which a high voltage is applied to a liquid to create an aerosol. It is especially useful in producing ions from macromolecules because it overcomes the propensity of these molecules to fragment when ionized. ESI is different from other ionization processes since it may produce multiple-charged ions, effectively extending the mass range of the analyser to accommodate the kDa-MDa orders of magnitude observed in proteins and their associated polypeptide fragments.
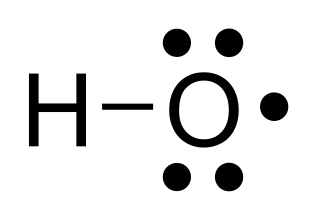
The hydroxyl radical is the diatomic molecule •
OH. The hydroxyl radical is very stable as a dilute gas, but it decays very rapidly in the condensed phase. It is pervasive in some situations. Most notably the hydroxyl radicals are produced from the decomposition of hydroperoxides (ROOH) or, in atmospheric chemistry, by the reaction of excited atomic oxygen with water. It is also important in the field of radiation chemistry, since it leads to the formation of hydrogen peroxide and oxygen, which can enhance corrosion and SCC in coolant systems subjected to radioactive environments. The unpaired electron of the hydroxyl radical is officially represented by a middle dot, •, beside the O.

In mass spectrometry, matrix-assisted laser desorption/ionization (MALDI) is an ionization technique that uses a laser energy-absorbing matrix to create ions from large molecules with minimal fragmentation. It has been applied to the analysis of biomolecules and various organic molecules, which tend to be fragile and fragment when ionized by more conventional ionization methods. It is similar in character to electrospray ionization (ESI) in that both techniques are relatively soft ways of obtaining ions of large molecules in the gas phase, though MALDI typically produces far fewer multi-charged ions.

Liquid chromatography–mass spectrometry (LC–MS) is an analytical chemistry technique that combines the physical separation capabilities of liquid chromatography with the mass analysis capabilities of mass spectrometry (MS). Coupled chromatography – MS systems are popular in chemical analysis because the individual capabilities of each technique are enhanced synergistically. While liquid chromatography separates mixtures with multiple components, mass spectrometry provides spectral information that may help to identify each separated component. MS is not only sensitive, but provides selective detection, relieving the need for complete chromatographic separation. LC–MS is also appropriate for metabolomics because of its good coverage of a wide range of chemicals. This tandem technique can be used to analyze biochemical, organic, and inorganic compounds commonly found in complex samples of environmental and biological origin. Therefore, LC–MS may be applied in a wide range of sectors including biotechnology, environment monitoring, food processing, and pharmaceutical, agrochemical, and cosmetic industries. Since the early 2000s, LC–MS has also begun to be used in clinical applications.

Atmospheric pressure chemical ionization (APCI) is an ionization method used in mass spectrometry which utilizes gas-phase ion-molecule reactions at atmospheric pressure (105 Pa), commonly coupled with high-performance liquid chromatography (HPLC). APCI is a soft ionization method similar to chemical ionization where primary ions are produced on a solvent spray. The main usage of APCI is for polar and relatively less polar thermally stable compounds with molecular weight less than 1500 Da. The application of APCI with HPLC has gained a large popularity in trace analysis detection such as steroids, pesticides and also in pharmacology for drug metabolites.
Hydrogen–deuterium exchange is a chemical reaction in which a covalently bonded hydrogen atom is replaced by a deuterium atom, or vice versa. It can be applied most easily to exchangeable protons and deuterons, where such a transformation occurs in the presence of a suitable deuterium source, without any catalyst. The use of acid, base or metal catalysts, coupled with conditions of increased temperature and pressure, can facilitate the exchange of non-exchangeable hydrogen atoms, so long as the substrate is robust to the conditions and reagents employed. This often results in perdeuteration: hydrogen-deuterium exchange of all non-exchangeable hydrogen atoms in a molecule.

Electron-transfer dissociation (ETD) is a method of fragmenting multiply-charged gaseous macromolecules in a mass spectrometer between the stages of tandem mass spectrometry (MS/MS). Similar to electron-capture dissociation, ETD induces fragmentation of large, multiply-charged cations by transferring electrons to them. ETD is used extensively with polymers and biological molecules such as proteins and peptides for sequence analysis. Transferring an electron causes peptide backbone cleavage into c- and z-ions while leaving labile post translational modifications (PTM) intact. The technique only works well for higher charge state peptide or polymer ions (z>2). However, relative to collision-induced dissociation (CID), ETD is advantageous for the fragmentation of longer peptides or even entire proteins. This makes the technique important for top-down proteomics. The method was developed by Hunt and coworkers at the University of Virginia.

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important method for the accurate mass determination and characterization of proteins, and a variety of methods and instrumentations have been developed for its many uses. Its applications include the identification of proteins and their post-translational modifications, the elucidation of protein complexes, their subunits and functional interactions, as well as the global measurement of proteins in proteomics. It can also be used to localize proteins to the various organelles, and determine the interactions between different proteins as well as with membrane lipids.

Desorption electrospray ionization (DESI) is an ambient ionization technique that can be coupled to mass spectrometry (MS) for chemical analysis of samples at atmospheric conditions. Coupled ionization sources-MS systems are popular in chemical analysis because the individual capabilities of various sources combined with different MS systems allow for chemical determinations of samples. DESI employs a fast-moving charged solvent stream, at an angle relative to the sample surface, to extract analytes from the surfaces and propel the secondary ions toward the mass analyzer. This tandem technique can be used to analyze forensics analyses, pharmaceuticals, plant tissues, fruits, intact biological tissues, enzyme-substrate complexes, metabolites and polymers. Therefore, DESI-MS may be applied in a wide variety of sectors including food and drug administration, pharmaceuticals, environmental monitoring, and biotechnology.
Michael L. Gross is Professor of Chemistry, Medicine, and Immunology, at Washington University in St. Louis. He was formerly Professor of Chemistry at the University of Nebraska-Lincoln from 1968–1994. He is recognized for his contributions to the field of mass spectrometry and ion chemistry. He is credited with the discovery of distonic ions, chemical species containing a radical and an ionic site on different atoms of the same molecule.

Ion mobility spectrometry–mass spectrometry (IMS-MS) is an analytical chemistry method that separates gas phase ions based on their interaction with a collision gas and their masses. In the first step, the ions are separated according to their mobility through a buffer gas on a millisecond timescale using an ion mobility spectrometer. The separated ions are then introduced into a mass analyzer in a second step where their mass-to-charge ratios can be determined on a microsecond timescale. The effective separation of analytes achieved with this method makes it widely applicable in the analysis of complex samples such as in proteomics and metabolomics.

Ambient ionization is a form of ionization in which ions are formed in an ion source outside the mass spectrometer without sample preparation or separation. Ions can be formed by extraction into charged electrospray droplets, thermally desorbed and ionized by chemical ionization, or laser desorbed or ablated and post-ionized before they enter the mass spectrometer.

In mass spectrometry, fragmentation is the dissociation of energetically unstable molecular ions formed from passing the molecules mass spectrum. These reactions are well documented over the decades and fragmentation patterns are useful to determine the molar weight and structural information of unknown molecules. Fragmentation that occurs in tandem mass spectrometry experiments has been a recent focus of research, because this data helps facilitate the identification of molecules.
Kevin Downard is a British - Australian academic scientist whose research specialises in the improving responses to infectious disease through the application and development of mass spectrometry and other molecular approaches in the life and medical sciences. Downard has over 35 years of experience in the field and has written over 145 lead-author scientific peer-reviewed journal publications, and two books including a textbook for the Royal Society of Chemistry and the first book to be published on the role of mass spectrometry in the study of protein interactions.
Free radical damage to DNA can occur as a result of exposure to ionizing radiation or to radiomimetic compounds. Damage to DNA as a result of free radical attack is called indirect DNA damage because the radicals formed can diffuse throughout the body and affect other organs. Malignant melanoma can be caused by indirect DNA damage because it is found in parts of the body not exposed to sunlight. DNA is vulnerable to radical attack because of the very labile hydrogens that can be abstracted and the prevalence of double bonds in the DNA bases that free radicals can easily add to.
Time-resolved mass spectrometry (TRMS) is a strategy in analytical chemistry that uses mass spectrometry platform to collect data with temporal resolution. Implementation of TRMS builds on the ability of mass spectrometers to process ions within sub-second duty cycles. It often requires the use of customized experimental setups. However, they can normally incorporate commercial mass spectrometers. As a concept in analytical chemistry, TRMS encompasses instrumental developments, methodological developments, and applications.

Atmospheric pressure photoionization (APPI) is a soft ionization method used in mass spectrometry (MS) usually coupled to liquid chromatography (LC). Molecules are ionized using a vacuum ultraviolet (VUV) light source operating at atmospheric pressure, either by direct absorption followed by electron ejection or through ionization of a dopant molecule that leads to chemical ionization of target molecules. The sample is usually a solvent spray that is vaporized by nebulization and heat. The benefit of APPI is that it ionizes molecules across a broad range of polarity and is particularly useful for ionization of low polarity molecules for which other popular ionization methods such as electrospray ionization (ESI) and atmospheric pressure chemical ionization (APCI) are less suitable. It is also less prone to ion suppression and matrix effects compared to ESI and APCI and typically has a wide linear dynamic range. The application of APPI with LC/MS is commonly used for analysis of petroleum compounds, pesticides, steroids, and drug metabolites lacking polar functional groups and is being extensively deployed for ambient ionization particularly for explosives detection in security applications.
Probe electrospray ionization (PESI) is an electrospray-based ambient ionization technique which is coupled with mass spectrometry for sample analysis. Unlike traditional mass spectrometry ion sources which must be maintained in a vacuum, ambient ionization techniques permit sample ionization under ambient conditions, allowing for the high-throughput analysis of samples in their native state, often with minimal or no sample pre-treatment. The PESI ion source simply consists of a needle to which a high voltage is applied following sample pick-up, initiating electrospray directly from the solid needle.
References
- ↑ Sheshberadaran H, Payne LG (January 1988). "Protein antigen-monoclonal antibody contact sites investigated by limited proteolysis of monoclonal antibody-bound antigen: protein "footprinting"". Proceedings of the National Academy of Sciences of the United States of America. 85 (1): 1–5. Bibcode:1988PNAS...85....1S. doi: 10.1073/pnas.85.1.1 . PMC 279469 . PMID 2448767.
- ↑ Shafer GE, Price MA, Tullius TD (1989). "Use of the hydroxyl radical and gel electrophoresis to study DNA structure". Electrophoresis. 10 (5–6): 397–404. doi:10.1002/elps.1150100518. PMID 2504579. S2CID 38355953.
- ↑ Galas DJ (November 2001). "The invention of footprinting". Trends in Biochemical Sciences. 26 (11): 690–3. doi:10.1016/S0968-0004(01)01979-X. PMID 11701330.
- ↑ Maleknia SD, Downard KM (May 2014). "Advances in radical probe mass spectrometry for protein footprinting in chemical biology applications". Chemical Society Reviews. 43 (10): 3244–58. doi:10.1039/C3CS60432B. PMID 24590115.
- ↑ Wang L, Chance MR (October 2011). "Structural mass spectrometry of proteins using hydroxyl radical based protein footprinting". Analytical Chemistry. 83 (19): 7234–41. doi:10.1021/ac200567u. PMC 3184339 . PMID 21770468.
- ↑ Downard KM, Maleknia SD (2019). "Mass spectrometry in structural proteomics: The case for radical probe protein footprinting". Trends in Analytical Chemistry. 110: 293–302. doi:10.1016/j.trac.2018.11.016.
- ↑ Maleknia SD, Brenowitz M, Chance MR (September 1999). "Millisecond radiolytic modification of peptides by synchrotron X-rays identified by mass spectrometry". Analytical Chemistry. 71 (18): 3965–73. doi:10.1021/ac990500e. PMID 10500483.
- ↑ Maleknia SD, Ralston CY, Brenowitz MD, Downard KM, Chance MR (February 2001). "Determination of macromolecular folding and structure by synchrotron x-ray radiolysis techniques". Analytical Biochemistry. 289 (2): 103–15. doi:10.1006/abio.2000.4910. PMID 11161303.
- ↑ Maleknia SD, Chance MR, Downard KM (1999). "Electrospray-assisted modification of proteins: a radical probe of protein structure". Rapid Communications in Mass Spectrometry. 13 (23): 2352–8. Bibcode:1999RCMS...13.2352M. doi:10.1002/(SICI)1097-0231(19991215)13:23<2352::AID-RCM798>3.0.CO;2-X. PMID 10567934.
- ↑ Hambly DM, Gross ML (December 2005). "Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale". Journal of the American Society for Mass Spectrometry. 16 (12): 2057–63. doi: 10.1016/j.jasms.2005.09.008 . PMID 16263307. S2CID 24995091.
- ↑ Gau BC, Sharp JS, Rempel DL, Gross ML (August 2009). "Fast photochemical oxidation of protein footprints faster than protein unfolding". Analytical Chemistry. 81 (16): 6563–71. doi:10.1021/ac901054w. PMC 3164994 . PMID 20337372.
- ↑ Vahidi S, Konermann L (2016). "Probing the time scale of FPOP (fast photochemical oxidation of proteins): radical reactions extend over tens of milliseconds". Journal of the American Society for Mass Spectrometry. 27 (7): 1156–1164. Bibcode:2016JASMS..27.1156V. doi:10.1007/s13361-016-1389-x. PMID 27067899. S2CID 35039048.
- ↑ Maleknia SD, Kiselar JG, Downard KM (2002). "Hydroxyl radical probe of the surface of lysozyme by synchrotron radiolysis and mass spectrometry". Rapid Communications in Mass Spectrometry. 16 (1): 53–61. Bibcode:2002RCMS...16...53M. doi:10.1002/rcm.543. PMID 11754247.
- ↑ Maleknia SD, Downard K (2001). "Radical approaches to probe protein structure, folding, and interactions by mass spectrometry". Mass Spectrometry Reviews. 20 (6): 388–401. Bibcode:2001MSRv...20..388M. doi:10.1002/mas.10013. PMID 11997945.
- ↑ Downard K (24 August 2007). Mass Spectrometry of Protein Interactions. John Wiley & Sons. ISBN 978-0-470-14632-3 . Retrieved 14 September 2013.
- ↑ Wong JW, Maleknia SD, Downard KM (April 2003). "Study of the ribonuclease-S-protein-peptide complex using a radical probe and electrospray ionization mass spectrometry". Analytical Chemistry. 75 (7): 1557–63. doi:10.1021/ac026400h. PMID 12705585.
- ↑ Gerega SK, Downard KM (July 2006). "PROXIMO--a new docking algorithm to model protein complexes using data from radical probe mass spectrometry (RP-MS)". Bioinformatics. 22 (14): 1702–9. doi: 10.1093/bioinformatics/btl178 . PMID 16679333.
- ↑ Downard KM, Kokabu Y, Ikeguchi M, Akashi S (November 2011). "Homology-modelled structure of the βB2B3-crystallin heterodimer studied by ion mobility and radical probe MS". The FEBS Journal. 278 (21): 4044–54. doi: 10.1111/j.1742-4658.2011.08309.x . PMID 21848669.
- ↑ Downard KM, Maleknia SD, Akashi S (February 2012). "Impact of limited oxidation on protein ion mobility and structure of importance to footprinting by radical probe mass spectrometry". Rapid Communications in Mass Spectrometry. 26 (3): 226–30. Bibcode:2012RCMS...26..226D. doi:10.1002/rcm.5320. PMID 22223306.
- ↑ Shum WK, Maleknia SD, Downard KM (September 2005). "Onset of oxidative damage in alpha-crystallin by radical probe mass spectrometry". Analytical Biochemistry. 344 (2): 247–56. doi:10.1016/j.ab.2005.06.035. PMID 16091281.