
Pseudomonas aeruginosa is a common encapsulated, Gram-negative, rod-shaped bacterium that can cause disease in plants and animals, including humans. A species of considerable medical importance, P. aeruginosa is a multidrug resistant pathogen recognized for its ubiquity, its intrinsically advanced antibiotic resistance mechanisms, and its association with serious illnesses – hospital-acquired infections such as ventilator-associated pneumonia and various sepsis syndromes.
The gene rpoS encodes the sigma factor sigma-38, a 37.8 kD protein in Escherichia coli. Sigma factors are proteins that regulate transcription in bacteria. Sigma factors can be activated in response to different environmental conditions. rpoS is transcribed in late exponential phase, and RpoS is the primary regulator of stationary phase genes. RpoS is a central regulator of the general stress response and operates in both a retroactive and a proactive manner: it not only allows the cell to survive environmental challenges, but it also prepares the cell for subsequent stresses (cross-protection). The transcriptional regulator CsgD is central to biofilm formation, controlling the expression of the curli structural and export proteins, and the diguanylate cyclase, adrA, which indirectly activates cellulose production. The rpoS gene most likely originated in the gammaproteobacteria.

The PrrF RNAs are small non-coding RNAs involved in iron homeostasis and are encoded by all Pseudomonas species. The PrrF RNAs are analogs of the RyhB RNA, which is encoded by enteric bacteria. Expression of the PrrF RNAs is repressed by the ferric uptake regulator (Fur) when cells are grown in iron-replete conditions. Under iron limitation, the PrrF RNAs are expressed and act to negatively regulate several genes encoding iron-containing proteins, including SodB and succinate dehydrogenase. As such, PrrF regulation "spares" iron when this nutrient becomes scarce.

The rsmY RNA family is a set of related non-coding RNA genes, that like RsmZ, is regulated by the GacS/GacA signal transduction system in the plant-beneficial soil bacterium and biocontrol model organism Pseudomonas fluorescens CHA0. GacA/GacS target genes are translationally repressed by the small RNA binding protein RsmA. RsmY and RsmZ RNAs bind RsmA to relieve this repression and so enhance secondary metabolism and biocontrol traits.

In molecular biology the ArcZ RNA is a small non-coding RNA (ncRNA). It is the functional product of a gene which is not translated into protein. ArcZ is an Hfq binding RNA that functions as an antisense regulator of a number of protein coding genes.

Pseudomonas sRNA P1 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis.
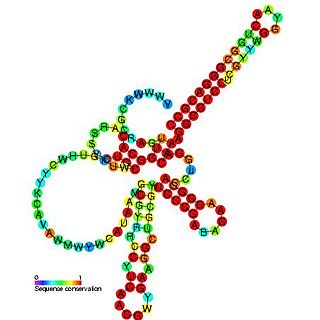
Pseudomonas sRNA P11 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis. P11 is located between a putative threonine protein kinase and putative nitrate reductase and is conserved in several Pseudomonas species. P11 has a predicted Rho independent terminator at the 3′ end but the function of P11 is unknown.
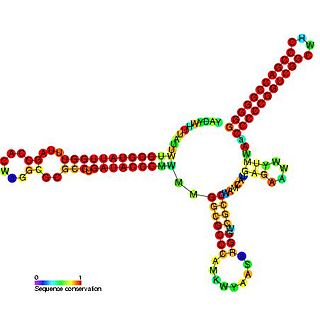
Pseudomonas sRNA P15 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis.
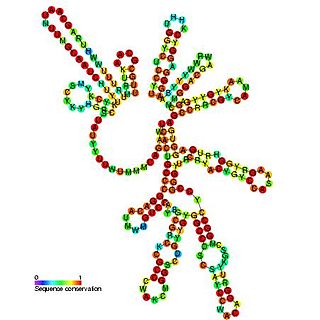
Pseudomonas sRNA P24 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis.
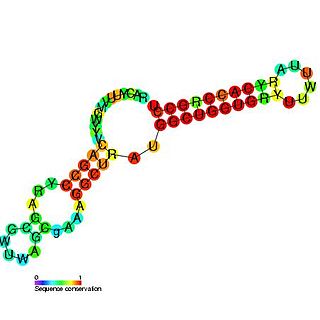
Pseudomonas sRNA P26 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis. P26 is conserved across many Gammaproteobacteria species and appears to be consistently located between the DNA directed RNA polymerase and 50S ribosomal protein L7/L12 genes.

Pseudomonas sRNA P9 is a ncRNA that was predicted using bioinformatic tools in the genome of the opportunistic pathogen Pseudomonas aeruginosa and its expression verified by northern blot analysis.
Pseudomonas sRNA are non-coding RNAs (ncRNA) that were predicted by the bioinformatic program SRNApredict2. This program identifies putative sRNAs by searching for co-localization of genetic features commonly associated with sRNA-encoding genes and the expression of the predicted sRNAs was subsequently confirmed by Northern blot analysis. These sRNAs have been shown to be conserved across several pseudomonas species but their function is yet to be determined. Using Tet-Trap genetic approach RNAT genes post-transcriptionally regulated by temperature upshift were identified: ptxS and PA5194.

The gyrA RNA motif is a conserved RNA structure identified by bioinformatics. The RNAs are present in multiple species of bacteria within the order Pseudomonadales. This order contains the genus Pseudomonas, which includes the opportunistic human pathogen Pseudomonas aeruginosa and Pseudomonas syringae, a plant pathogen.

The Pseudomon-1 RNA motif is a conserved RNA identified by bioinformatics. It is used by most species whose genomes have been sequenced and that are classified within the genus Pseudomonas, and is also present in Azotobacter vinelandii, a closely related species. It is presumed to function as a non-coding RNA. Pseudomon-1 RNAs consistently have a downstream rho-independent transcription terminator.
Bacterial small RNAs (sRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.

Mycobacterium tuberculosis contains at least nine small RNA families in its genome. The small RNA (sRNA) families were identified through RNomics – the direct analysis of RNA molecules isolated from cultures of Mycobacterium tuberculosis. The sRNAs were characterised through RACE mapping and Northern blot experiments. Secondary structures of the sRNAs were predicted using Mfold.
αr9 is a family of bacterial small non-coding RNAs with representatives in a broad group of α-proteobacteria from the order Rhizobiales. The first member of this family (Smr9C) was found in a Sinorhizobium meliloti 1021 locus located in the chromosome (C). Further homology and structure conservation analysis have identified full-length Smr9C homologs in several nitrogen-fixing symbiotic rhizobia, in the plant pathogens belonging to Agrobacterium species as well as in a broad spectrum of Brucella species. αr9C RNA species are 144-158 nt long and share a well defined common secondary structure consisting of seven conserved regions. Most of the αr9 transcripts can be catalogued as trans-acting sRNAs expressed from well-defined promoter regions of independent transcription units within intergenic regions (IGRs) of the α-proteobacterial genomes.
αr14 is a family of bacterial small non-coding RNAs with representatives in a broad group of α-proteobacteria. The first member of this family (Smr14C2) was found in a Sinorhizobium meliloti 1021 locus located in the chromosome (C). It was later renamed NfeR1 and shown to be highly expressed in salt stress and during the symbiotic interaction on legume roots. Further homology and structure conservation analysis identified 2 other chromosomal copies and 3 plasmidic ones. Moreover, full-length Smr14C homologs have been identified in several nitrogen-fixing symbiotic rhizobia, in the plant pathogens belonging to Agrobacterium species as well as in a broad spectrum of Brucella species. αr14C RNA species are 115-125 nt long and share a well defined common secondary structure. Most of the αr14 transcripts can be catalogued as trans-acting sRNAs expressed from well-defined promoter regions of independent transcription units within intergenic regions (IGRs) of the α-proteobacterial genomes.
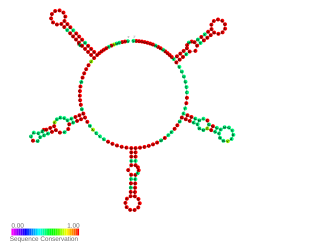
NrsZ is a bacterial small RNA found in the opportunistic pathogen Pseudomonas aeruginosa PAO1. Its transcription is induced during nitrogen limitation by the NtrB/C two-component system together with the alternative sigma factor RpoN. NrsZ by activating rhlA positively regulates the production of rhamnolipid surfactants needed for swarming motility.
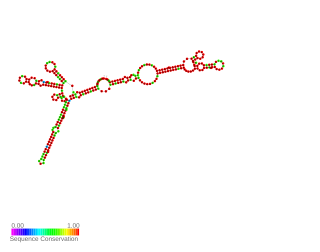
AsponA is a small asRNA transcribed antisense to the penicillin-binding protein 1A gene called ponA. It was identified by RNAseq and the expression was validated by 5' and 3' RACE experiments in Pseudomanas aeruginosa. AsponA expression was up or down regulated under different antibiotic stress. Due to it s location it may be able to prevent the transcription or translation of the opposite gene. Study by Wurtzel et al. and Ferrara et al. also detected its expression.















