In genetics, attenuation is a proposed mechanism of control in some bacterial operons which results in premature termination of transcription and is based on the fact that, in bacteria, transcription and translation proceed simultaneously. Attenuation involves a provisional stop signal (attenuator), located in the DNA segment that corresponds to the leader sequence of mRNA. During attenuation, the ribosome becomes stalled (delayed) in the attenuator region in the mRNA leader. Depending on the metabolic conditions, the attenuator either stops transcription at that point or allows read-through to the structural gene part of the mRNA and synthesis of the appropriate protein.

Intrinsic, or rho-independent termination, is a process in prokaryotes to signal the end of transcription and release the newly constructed RNA molecule. In prokaryotes such as E. coli, transcription is terminated either by a rho-dependent process or rho-independent process. In the Rho-dependent process, the rho-protein locates and binds the signal sequence in the mRNA and signals for cleavage. Contrarily, intrinsic termination does not require a special protein to signal for termination and is controlled by the specific sequences of RNA. When the termination process begins, the transcribed mRNA forms a stable secondary structure hairpin loop, also known as a Stem-loop. This RNA hairpin is followed by multiple uracil nucleotides. The bonds between uracil and adenine are very weak. A protein bound to RNA polymerase (nusA) binds to the stem-loop structure tightly enough to cause the polymerase to temporarily stall. This pausing of the polymerase coincides with transcription of the poly-uracil sequence. The weak adenine-uracil bonds lower the energy of destabilization for the RNA-DNA duplex, allowing it to unwind and dissociate from the RNA polymerase. Overall, the modified RNA structure is what terminates transcription.

The msiK RNA motif describes a conserved RNA structure discovered using bioinformatics. The RNA is always found in the presumed 5' untranslated regions of genes annotated as msiK, and is therefore hypothesized to be an RNA-based cis-regulatory element that regulates these genes.

The eps-Associated RNA element is a conserved RNA motif associated with exopolysaccharide (eps) or capsule biosynthesis genes in a subset of bacteria classified within the order Bacillales. It was initially discovered in Bacillus subtilis, located between the second and third gene in the eps operon. Deletion of the EAR element impairs biofilm formation.

The Actino-ugpB RNA motif is a conserved RNA structure that was discovered by bioinformatics. Actino-ugpB motifs are found in strains of the species Gardnerella vaginalis, within the phylum Actinobacteria.

The che1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. che1 motifs are found in the genus Streptomyces, as well as some other organisms that are closely related to this genus.

The D12-methyl RNA motif is a conserved RNA structure that was discovered by bioinformatics. D12-methyl motifs are found in metagenomic DNA samples, and have not yet been found in a classified organism.

The dfrA-dnaX RNA motif is a conserved RNA structure that was discovered by bioinformatics. dfrA-dnaX motifs are found in Parvimonas.

The DUF2800 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF2800 motif RNAs are found in Firmicutes. DUF2800 RNAs are also predicted in the phyla Actinobacteria and Synergistetes, although these RNAs are likely the result of recent horizontal gene transfer or conceivably sequence contamination.

The DUF3577 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF3577 motifs are found in the organism Cardiobacterium valvarum and metagenomic sequences from unknown organisms.

The DUF805 RNA motif is a conserved RNA structure that was discovered by bioinformatics. The motif is subdivided into the DUF805 motif and the DUF805b motif, which have similar, but distinct secondary structures. Together, these motifs are found in Bacteroidetes, Chlorobi AND Proteobacteria.

The ftsZ-DE RNA motif is a conserved RNA structure that was discovered by bioinformatics. ftsZ-DE motifs are found in bacteria belonging to the genus Fibrobacter.

The FuFi-1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Such RNA "motifs" are often the first step to elucidating the biological function of a novel RNA. FuFi-1 motif RNAs are found in Firmicutes AND Fusobacteria.

The gntR-DTE RNA motif is a conserved RNA structure that was discovered by bioinformatics. gntR-DTE motifs are found in some, but not all species within the genus Streptomyces.

The Latescibacteria, OD1, OP11, TM7 RNA motif is a conserved RNA structure that was discovered by bioinformatics. LOOT motif RNAs are found in multiple bacterial phyla that have only recently been discovered, and are currently not well understood: Latescibacteria, OD1/Parcubacteria, OP11 AND TM7. In some cases, no specific organism has been isolated in the relevant phylum, but the existence of the bacterial phylum is known only through analysis of metagenomic sequences. Curiously, the LOOT motif is not known in any phylum that has been studied for a long time.

The NLPC-P60 RNA motif is a conserved RNA structure that was discovered by bioinformatics. NLPC-P60 motif RNAs are found in Streptomyces.
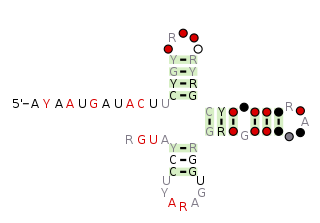
The pemK RNA motif is a conserved RNA structure that was discovered by bioinformatics. pemK motif RNAs are found in organisms within the phylum Firmicutes, and is very widespread in this phylum.

The raiA RNA motif is a conserved RNA structure that was discovered by bioinformatics. raiA motif RNAs are found in Actinobacteria AND Firmicutes, and have many conserved features—including conserved nucleotide positions, conserved secondary structures and associated protein-coding genes—in both of these phyla. Some conserved features of the raiA RNA motif suggest that they function as cis-regulatory elements, but other aspects of the motif suggest otherwise.
The Streptomyces-atpC RNA motif is a conserved RNA structure that was discovered by bioinformatics. Streptomyces-atpC motif RNAs are found in organisms classified within the genus Streptomyces.

The uup RNA motif is a conserved RNA structure that was discovered by bioinformatics. uup motif RNAs are found in Firmicutes and Gammaproteobacteria.


















