Example
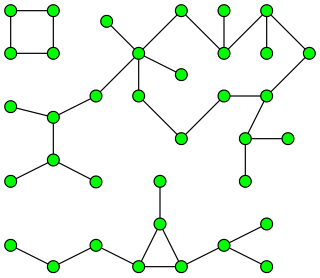
In the following example green edges are being evaluated by the algorithm and red edges have been deleted.
The reverse-delete algorithm is an algorithm in graph theory used to obtain a minimum spanning tree from a given connected, edge-weighted graph. It first appeared in Kruskal (1956), but it should not be confused with Kruskal's algorithm which appears in the same paper. If the graph is disconnected, this algorithm will find a minimum spanning tree for each disconnected part of the graph. The set of these minimum spanning trees is called a minimum spanning forest, which contains every vertex in the graph.
This algorithm is a greedy algorithm, choosing the best choice given any situation. It is the reverse of Kruskal's algorithm, which is another greedy algorithm to find a minimum spanning tree. Kruskal’s algorithm starts with an empty graph and adds edges while the Reverse-Delete algorithm starts with the original graph and deletes edges from it. The algorithm works as follows:
function ReverseDelete(edges[] E) issortE in decreasing order Define an index i ← 0 whilei < size(E) do Define edge ← E[i] deleteE[i] if graph is not connected thenE[i] ← edgei ← i + 1 return edges[] E
In the above the graph is the set of edges E with each edge containing a weight and connected vertices v1 and v2.
In the following example green edges are being evaluated by the algorithm and red edges have been deleted.
The algorithm can be shown to run in O(E log V (log log V)3) time (using big-O notation), where E is the number of edges and V is the number of vertices. This bound is achieved as follows:
It is recommended to read the proof of the Kruskal's algorithm first.
The proof consists of two parts. First, it is proved that the edges that remain after the algorithm is applied form a spanning tree. Second, it is proved that the spanning tree is of minimal weight.
The remaining sub-graph (g) produced by the algorithm is not disconnected since the algorithm checks for that in line 7. The result sub-graph cannot contain a cycle since if it does then when moving along the edges we would encounter the max edge in the cycle and we would delete that edge. Thus g must be a spanning tree of the main graph G.
We show that the following proposition P is true by induction: If F is the set of edges remained at the end of the while loop, then there is some minimum spanning tree that (its edges) are a subset of F.

A minimum spanning tree (MST) or minimum weight spanning tree is a subset of the edges of a connected, edge-weighted undirected graph that connects all the vertices together, without any cycles and with the minimum possible total edge weight. That is, it is a spanning tree whose sum of edge weights is as small as possible. More generally, any edge-weighted undirected graph has a minimum spanning forest, which is a union of the minimum spanning trees for its connected components.

Kruskal's algorithm finds a minimum spanning forest of an undirected edge-weighted graph. If the graph is connected, it finds a minimum spanning tree. It is a greedy algorithm that in each step adds to the forest the lowest-weight edge that will not form a cycle. The key steps of the algorithm are sorting and the use of a disjoint-set data structure to detect cycles. Its running time is dominated by the time to sort all of the graph edges by their weight.

In computer science, Prim's algorithm is a greedy algorithm that finds a minimum spanning tree for a weighted undirected graph. This means it finds a subset of the edges that forms a tree that includes every vertex, where the total weight of all the edges in the tree is minimized. The algorithm operates by building this tree one vertex at a time, from an arbitrary starting vertex, at each step adding the cheapest possible connection from the tree to another vertex.

Borůvka's algorithm is a greedy algorithm for finding a minimum spanning tree in a graph, or a minimum spanning forest in the case of a graph that is not connected.
This is a glossary of graph theory. Graph theory is the study of graphs, systems of nodes or vertices connected in pairs by lines or edges.
In graph theory, the Robertson–Seymour theorem states that the undirected graphs, partially ordered by the graph minor relationship, form a well-quasi-ordering. Equivalently, every family of graphs that is closed under minors can be defined by a finite set of forbidden minors, in the same way that Wagner's theorem characterizes the planar graphs as being the graphs that do not have the complete graph K5 or the complete bipartite graph K3,3 as minors.

In the mathematical field of graph theory, a spanning treeT of an undirected graph G is a subgraph that is a tree which includes all of the vertices of G. In general, a graph may have several spanning trees, but a graph that is not connected will not contain a spanning tree. If all of the edges of G are also edges of a spanning tree T of G, then G is a tree and is identical to T.
In the mathematical theory of matroids, a graphic matroid is a matroid whose independent sets are the forests in a given finite undirected graph. The dual matroids of graphic matroids are called co-graphic matroids or bond matroids. A matroid that is both graphic and co-graphic is sometimes called a planar matroid ; these are exactly the graphic matroids formed from planar graphs.

A Euclidean minimum spanning tree of a finite set of points in the Euclidean plane or higher-dimensional Euclidean space connects the points by a system of line segments with the points as endpoints, minimizing the total length of the segments. In it, any two points can reach each other along a path through the line segments. It can be found as the minimum spanning tree of a complete graph with the points as vertices and the Euclidean distances between points as edge weights.
In graph theory, a cut is a partition of the vertices of a graph into two disjoint subsets. Any cut determines a cut-set, the set of edges that have one endpoint in each subset of the partition. These edges are said to cross the cut. In a connected graph, each cut-set determines a unique cut, and in some cases cuts are identified with their cut-sets rather than with their vertex partitions.
The Christofides algorithm or Christofides–Serdyukov algorithm is an algorithm for finding approximate solutions to the travelling salesman problem, on instances where the distances form a metric space . It is an approximation algorithm that guarantees that its solutions will be within a factor of 3/2 of the optimal solution length, and is named after Nicos Christofides and Anatoliy I. Serdyukov ; the latter discovered it independently in 1976.

The distributed minimum spanning tree (MST) problem involves the construction of a minimum spanning tree by a distributed algorithm, in a network where nodes communicate by message passing. It is radically different from the classical sequential problem, although the most basic approach resembles Borůvka's algorithm. One important application of this problem is to find a tree that can be used for broadcasting. In particular, if the cost for a message to pass through an edge in a graph is significant, an MST can minimize the total cost for a source process to communicate with all the other processes in the network.
In graph theory, Edmonds' algorithm or Chu–Liu/Edmonds' algorithm is an algorithm for finding a spanning arborescence of minimum weight . It is the directed analog of the minimum spanning tree problem. The algorithm was proposed independently first by Yoeng-Jin Chu and Tseng-Hong Liu (1965) and then by Jack Edmonds (1967).
In graph theory, a pseudoforest is an undirected graph in which every connected component has at most one cycle. That is, it is a system of vertices and edges connecting pairs of vertices, such that no two cycles of consecutive edges share any vertex with each other, nor can any two cycles be connected to each other by a path of consecutive edges. A pseudotree is a connected pseudoforest.

A top tree is a data structure based on a binary tree for unrooted dynamic trees that is used mainly for various path-related operations. It allows simple divide-and-conquer algorithms. It has since been augmented to maintain dynamically various properties of a tree such as diameter, center and median.

In graph algorithms, the widest path problem is the problem of finding a path between two designated vertices in a weighted graph, maximizing the weight of the minimum-weight edge in the path. The widest path problem is also known as the maximum capacity path problem. It is possible to adapt most shortest path algorithms to compute widest paths, by modifying them to use the bottleneck distance instead of path length. However, in many cases even faster algorithms are possible.
The expected linear time MST algorithm is a randomized algorithm for computing the minimum spanning forest of a weighted graph with no isolated vertices. It was developed by David Karger, Philip Klein, and Robert Tarjan. The algorithm relies on techniques from Borůvka's algorithm along with an algorithm for verifying a minimum spanning tree in linear time. It combines the design paradigms of divide and conquer algorithms, greedy algorithms, and randomized algorithms to achieve expected linear performance.
In mathematics, a minimum bottleneck spanning tree (MBST) in an undirected graph is a spanning tree in which the most expensive edge is as cheap as possible. A bottleneck edge is the highest weighted edge in a spanning tree. A spanning tree is a minimum bottleneck spanning tree if the graph does not contain a spanning tree with a smaller bottleneck edge weight. For a directed graph, a similar problem is known as Minimum Bottleneck Spanning Arborescence (MBSA).
In computing and graph theory, a dynamic connectivity structure is a data structure that dynamically maintains information about the connected components of a graph.
In graph theory a minimum spanning tree (MST) of a graph with and is a tree subgraph of that contains all of its vertices and is of minimum weight.