Related Research Articles

Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair, which requires a homologous sequence to guide repair. The term "non-homologous end joining" was coined in 1996 by Moore and Haber.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids. It is widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses.
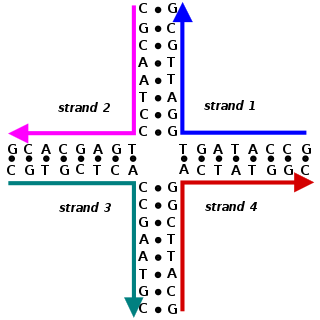
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the junction. The structure is named after Robin Holliday, the molecular biologist who proposed its existence in 1964.
Mitotic recombination is a type of genetic recombination that may occur in somatic cells during their preparation for mitosis in both sexual and asexual organisms. In asexual organisms, the study of mitotic recombination is one way to understand genetic linkage because it is the only source of recombination within an individual. Additionally, mitotic recombination can result in the expression of recessive genes in an otherwise heterozygous individual. This expression has important implications for the study of tumorigenesis and lethal recessive genes. Mitotic homologous recombination occurs mainly between sister chromatids subsequent to replication. Inter-sister homologous recombination is ordinarily genetically silent. During mitosis the incidence of recombination between non-sister homologous chromatids is only about 1% of that between sister chromatids.

Spo11 is a protein that in humans is encoded by the SPO11 gene. Spo11, in a complex with mTopVIB, creates double strand breaks to initiate meiotic recombination. Its active site contains a tyrosine which ligates and dissociates with DNA to promote break formation. One Spo11 protein is involved per strand of DNA, thus two Spo11 proteins are involved in each double stranded break event.
Chromosome segregation is the process in eukaryotes by which two sister chromatids formed as a consequence of DNA replication, or paired homologous chromosomes, separate from each other and migrate to opposite poles of the nucleus. This segregation process occurs during both mitosis and meiosis. Chromosome segregation also occurs in prokaryotes. However, in contrast to eukaryotic chromosome segregation, replication and segregation are not temporally separated. Instead segregation occurs progressively following replication.

Double-strand break repair protein MRE11 is an enzyme that in humans is encoded by the MRE11 gene. The gene has been designated MRE11A to distinguish it from the pseudogene MRE11B that is nowadays named MRE11P1.

DNA repair protein RAD50, also known as RAD50, is a protein that in humans is encoded by the RAD50 gene.

Exonuclease 1 is an enzyme that in humans is encoded by the EXO1 gene.

RAD52 homolog , also known as RAD52, is a protein which in humans is encoded by the RAD52 gene.

DNA repair and recombination protein RAD54-like is a protein that in humans is encoded by the RAD54L gene.

Crossover junction endonuclease MUS81 is an enzyme that in humans is encoded by the MUS81 gene.
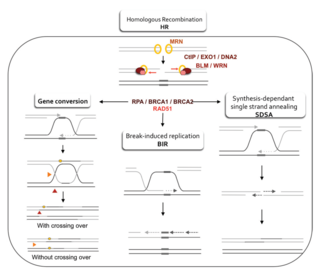
Homology directed repair (HDR) is a mechanism in cells to repair double-strand DNA lesions. The most common form of HDR is homologous recombination. The HDR mechanism can only be used by the cell when there is a homologous piece of DNA present in the nucleus, mostly in G2 and S phase of the cell cycle. Other examples of homology-directed repair include single-strand annealing and breakage-induced replication. When the homologous DNA is absent, another process called non-homologous end joining (NHEJ) takes place instead.
The MRX complex is a heterotrimeric protein complex consisting of Mre11, Rad50, and Xrs2. It is a budding yeast homolog of the mammalian Mre11-Rad50-Nbs1 (MRN) DNA damage repair complex.
The MRN complex is a protein complex consisting of Mre11, Rad50 and Nbs1. In eukaryotes, the MRN/X complex plays an important role in the initial processing of double-strand DNA breaks prior to repair by homologous recombination or non-homologous end joining. The MRN complex binds avidly to double-strand breaks both in vitro and in vivo and may serve to tether broken ends prior to repair by non-homologous end joining or to initiate DNA end resection prior to repair by homologous recombination. The MRN complex also participates in activating the checkpoint kinase ATM in response to DNA damage. Production of short single-strand oligonucleotides by Mre11 endonuclease activity has been implicated in ATM activation by the MRN complex.
Sgs1, also known as slow growth suppressor 1, is a DNA helicase protein found in Saccharomyces cerevisiae. It is a homolog of the bacterial RecQ helicase. Like the other members of the RecQ helicase family, Sgs1 is important for DNA repair. In particular, Sgs1 collaborates with other proteins to repair double-strand breaks during homologous recombination in eukaryotes.

Synthesis-dependent strand annealing (SDSA) is a major mechanism of homology-directed repair of DNA double-strand breaks (DSBs). Although many of the features of SDSA were first suggested in 1976, the double-Holliday junction model proposed in 1983 was favored by many researchers. In 1994, studies of double-strand gap repair in Drosophila were found to be incompatible with the double-Holliday junction model, leading researchers to propose a model they called synthesis-dependent strand annealing. Subsequent studies of meiotic recombination in S. cerevisiae found that non-crossover products appear earlier than double-Holliday junctions or crossover products, challenging the previous notion that both crossover and non-crossover products are produced by double-Holliday junctions and leading the authors to propose that non-crossover products are generated through SDSA.
Stephen Charles Kowalczykowski is a Distinguished Professor of Microbiology and Molecular Genetics at the University of California at Davis. His research focuses on the biochemistry and molecular biology of DNA repair and homologous recombination. His lab combines fluorescence microscopy, optical trapping and microfluidics to manipulate and visualize single molecules of DNA and the enzymes involved in processing and repairing DNA. He calls this scientific approach, "visual biochemistry". Stephen Kowalczykowski was elected to the American Society for Arts and Science in 2005, the National Academy of Sciences in 2007 and was a Harvey Society Lecturer at Rockefeller University in 2012.

DNA end resection, also called 5′–3′ degradation, is a biochemical process where the blunt end of a section of double-stranded DNA (dsDNA) is modified by cutting away some nucleotides from the 5' end to produce a 3' single-stranded sequence. The presence of a section of single-stranded DNA (ssDNA) allows the broken end of the DNA to line up accurately with a matching sequence, so that it can be accurately repaired.

A double-strand break repair model refers to the various models of pathways that cells undertake to repair double strand-breaks (DSB). DSB repair is an important cellular process, as the accumulation of unrepaired DSB could lead to chromosomal rearrangements, tumorigenesis or even cell death. In human cells, there are two main DSB repair mechanisms: Homologous recombination (HR) and non-homologous end joining (NHEJ). HR relies on undamaged template DNA as reference to repair the DSB, resulting in the restoration of the original sequence. NHEJ modifies and ligates the damaged ends regardless of homology. In terms of DSB repair pathway choice, most mammalian cells appear to favor NHEJ rather than HR. This is because the employment of HR may lead to gene deletion or amplification in cells which contains repetitive sequences. In terms of repair models in the cell cycle, HR is only possible during the S and G2 phases, while NHEJ can occur throughout whole process. These repair pathways are all regulated by the overarching DNA damage response mechanism. Besides HR and NHEJ, there are also other repair models which exists in cells. Some are categorized under HR, such as synthesis-dependent strain annealing, break-induced replication, and single-strand annealing; while others are an entirely alternate repair model, namely, the pathway microhomology-mediated end joining (MMEJ).
References
- ↑ Rattray, AJ; McGill, CB; Shafer, BK; Strathern, JN (May 2001). "Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1". Genetics. 158 (1): 109–22. doi:10.1093/genetics/158.1.109. PMC 1461648 . PMID 11333222.
- 1 2 3 4 Andres, Sara N.; Williams, R. Scott (August 2017). "CtIP/Ctp1/Sae2, molecular form fit for function". DNA Repair. 56: 109–117. doi:10.1016/j.dnarep.2017.06.013. PMC 5543718 . PMID 28623092.
- ↑ Huertas, Pablo; Cortés-Ledesma, Felipe; Sartori, Alessandro A.; Aguilera, Andrés; Jackson, Stephen P. (October 2008). "CDK targets Sae2 to control DNA-end resection and homologous recombination". Nature. 455 (7213): 689–692. Bibcode:2008Natur.455..689H. doi:10.1038/nature07215. PMC 2635538 . PMID 18716619.
- ↑ Clerici, Michela; Mantiero, Davide; Lucchini, Giovanna; Longhese, Maria Pia (18 November 2005). "The Saccharomyces cerevisiae Sae2 Protein Promotes Resection and Bridging of Double Strand Break Ends". Journal of Biological Chemistry. 280 (46): 38631–38638. doi: 10.1074/jbc.M508339200 . PMID 16162495.
- ↑ Lengsfeld, Bettina M.; Rattray, Alison J.; Bhaskara, Venugopal; Ghirlando, Rodolfo; Paull, Tanya T. (November 2007). "Sae2 Is an Endonuclease that Processes Hairpin DNA Cooperatively with the Mre11/Rad50/Xrs2 Complex". Molecular Cell. 28 (4): 638–651. doi:10.1016/j.molcel.2007.11.001. PMC 2194599 . PMID 18042458.