
Histone methyltransferases (HMT) are histone-modifying enzymes, that catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues of histone proteins. The attachment of methyl groups occurs predominantly at specific lysine or arginine residues on histones H3 and H4. Two major types of histone methyltranferases exist, lysine-specific and arginine-specific. In both types of histone methyltransferases, S-Adenosyl methionine (SAM) serves as a cofactor and methyl donor group.
The genomic DNA of eukaryotes associates with histones to form chromatin. The level of chromatin compaction depends heavily on histone methylation and other post-translational modifications of histones. Histone methylation is a principal epigenetic modification of chromatin that determines gene expression, genomic stability, stem cell maturation, cell lineage development, genetic imprinting, DNA methylation, and cell mitosis.
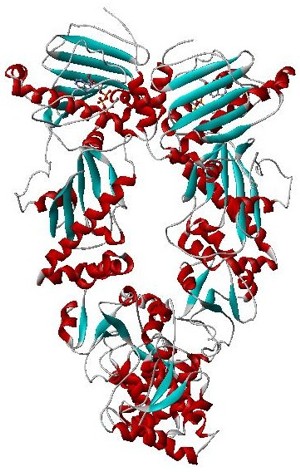
Hsp90 is a chaperone protein that assists other proteins to fold properly, stabilizes proteins against heat stress, and aids in protein degradation. It also stabilizes a number of proteins required for tumor growth, which is why Hsp90 inhibitors are investigated as anti-cancer drugs.
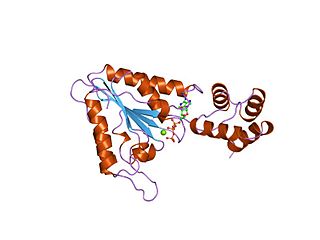
AAA proteins or ATPases Associated with diverse cellular Activities are a protein family sharing a common conserved module of approximately 230 amino acid residues. This is a large, functionally diverse protein family belonging to the AAA+ protein superfamily of ring-shaped P-loop NTPases, which exert their activity through the energy-dependent remodeling or translocation of macromolecules.
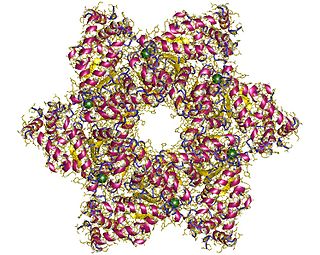
SV40 large T antigen is a hexamer protein that is a dominant-acting oncoprotein derived from the polyomavirus SV40. TAg is capable of inducing malignant transformation of a variety of cell types. The transforming activity of TAg is due in large part to its perturbation of the retinoblastoma (pRb) and p53 tumor suppressor proteins. In addition, TAg binds to several other cellular factors, including the transcriptional co-activators p300 and CBP, which may contribute to its transformation function. Similar proteins from related viruses are known as large tumor antigen in general.

The autoimmune regulator (AIRE) is a protein that in humans is encoded by the AIRE gene. It is a 13kb gene on chromosome 21q22.3 that has 545 amino acids. AIRE is a transcription factor expressed in the medulla of the thymus. It is part of the mechanism which eliminates self-reactive T cells that would cause autoimmune disease. It exposes T cells to normal, healthy proteins from all parts of the body, and T cells that react to those proteins are destroyed.
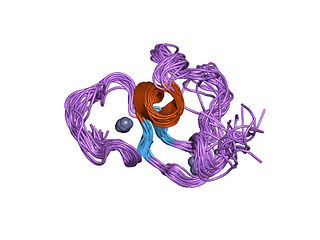
The PHD finger was discovered in 1993 as a Cys4-His-Cys3 motif in the plant homeodomain proteins HAT3.1 in Arabidopsis and maize ZmHox1a. The PHD zinc finger motif resembles the metal binding RING domain (Cys3-His-Cys4) and FYVE domain. It occurs as a single finger, but often in clusters of two or three, and it also occurs together with other domains, such as the chromodomain and the bromodomain.
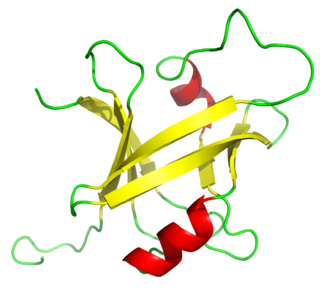
The K Homology (KH) domain is a protein domain that was first identified in the human heterogeneous nuclear ribonucleoprotein (hnRNP) K. An evolutionarily conserved sequence of around 70 amino acids, the KH domain is present in a wide variety of nucleic acid-binding proteins. The KH domain binds RNA, and can function in RNA recognition. It is found in multiple copies in several proteins, where they can function cooperatively or independently. For example, in the AU-rich element RNA-binding protein KSRP, which has 4 KH domains, KH domains 3 and 4 behave as independent binding modules to interact with different regions of the AU-rich RNA targets. The solution structure of the first KH domain of FMR1 and of the C-terminal KH domain of hnRNP K determined by nuclear magnetic resonance (NMR) revealed a beta-alpha-alpha-beta-beta-alpha structure. Autoantibodies to NOVA1, a KH domain protein, cause paraneoplastic opsoclonus ataxia. The KH domain is found at the N-terminus of the ribosomal protein S3. This domain is unusual in that it has a different fold compared to the normal KH domain.

In molecular biology, a RING (short for Really Interesting New Gene) finger domain is a protein structural domain of zinc finger type which contains a C3HC4 amino acid motif which binds two zinc cations (seven cysteines and one histidine arranged non-consecutively). This protein domain contains 40 to 60 amino acids. Many proteins containing a RING finger play a key role in the ubiquitination pathway.
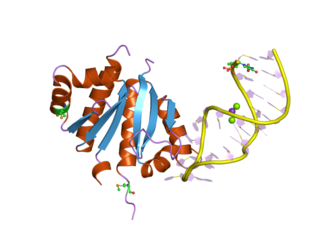
The B3 DNA binding domain (DBD) is a highly conserved domain found exclusively in transcription factors combined with other domains. It consists of 100-120 residues, includes seven beta strands and two alpha helices that form a DNA-binding pseudobarrel protein fold ; it interacts with the major groove of DNA.

S-Adenosylmethionine synthetase, also known as methionine adenosyltransferase (MAT), is an enzyme that creates S-adenosylmethionine by reacting methionine and ATP.

In molecular biology, the SAG1 protein domain is an example of a group of glycosylphosphatidylinositol (GPI)-linked proteins named SRSs. SAG1 is found on the surface of a protozoan parasite Toxoplasma gondii. This parasite infects almost any warm-blooded vertebrate. The surface of T. gondii is coated with a family of developmentally regulated glycosylphosphatidylinositol (GPI)-linked proteins (SRSs), of which SAG1 is the prototypic member.

The SQUAMOSA promoter binding protein-like family of transcription factors are defined by a plant-specific DNA-binding domain. The founding member of the family was identified based on its specific in vitro binding to the promoter of the snapdragon SQUAMOSA gene. SBP proteins are thought to be transcriptional activators.

In molecular biology the MYND-type zinc finger domain is a conserved protein domain. The MYND domain is present in a large group of proteins that includes RP-8 (PDCD2), Nervy, and predicted proteins from Drosophila, mammals, Caenorhabditis elegans, yeast, and plants. The MYND domain consists of a cluster of cysteine and histidine residues, arranged with an invariant spacing to form a potential zinc-binding motif. Mutating conserved cysteine residues in the DEAF-1 MYND domain does not abolish DNA binding, which suggests that the MYND domain might be involved in protein-protein interactions. Indeed, the MYND domain of ETO/MTG8 interacts directly with the N-CoR and SMRT co-repressors. Aberrant recruitment of co-repressor complexes and inappropriate transcriptional repression is believed to be a general mechanism of leukemogenesis caused by the t(8;21) translocations that fuse ETO with the acute myelogenous leukemia 1 (AML1) protein. ETO has been shown to be a co-repressor recruited by the promyelocytic leukemia zinc finger (PLZF) protein. A divergent MYND domain present in the adenovirus E1A binding protein BS69 was also shown to interact with N-CoR and mediate transcriptional repression. The current evidence suggests that the MYND motif in mammalian proteins constitutes a protein-protein interaction domain that functions as a co-repressor-recruiting interface.

In molecular biology the B-box-type zinc finger domain is a short protein domain of around 40 amino acid residues in length. B-box zinc fingers can be divided into two groups, where types 1 and 2 B-box domains differ in their consensus sequence and in the spacing of the 7-8 zinc-binding residues. Several proteins contain both types 1 and 2 B-boxes, suggesting some level of cooperativity between these two domains.
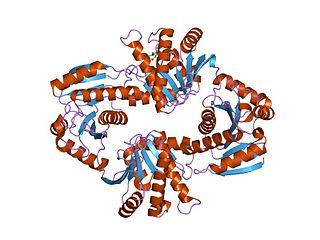
In molecular biology, the Macro domain or A1pp domain is a module of about 180 amino acids which can bind ADP-ribose, an NAD metabolite, or related ligands. Binding to ADP-ribose can be either covalent or non-covalent: in certain cases it is believed to bind non-covalently, while in other cases it appears to bind both non-covalently through a zinc finger motif, and covalently through a separate region of the protein.

In molecular biology, the GYF domain is an approximately 60-amino acid protein domain which contains a conserved GP[YF]xxxx[MV]xxWxxx[GN]YF motif. It was identified in the human intracellular protein termed CD2 binding protein 2 (CD2BP2), which binds to a site containing two tandem PPPGHR segments within the cytoplasmic region of CD2. Binding experiments and mutational analyses have demonstrated the critical importance of the GYF tripeptide in ligand binding. A GYF domain is also found in several other eukaryotic proteins of unknown function. It has been proposed that the GYF domain found in these proteins could also be involved in proline-rich sequence recognition. Resolution of the structure of the CD2BP2 GYF domain by NMR spectroscopy revealed a compact domain with a beta-beta-alpha-beta-beta topology, where the single alpha-helix is tilted away from the twisted, anti-parallel beta-sheet. The conserved residues of the GYF domain create a contiguous patch of predominantly hydrophobic nature which forms an integral part of the ligand-binding site. There is limited homology within the C-terminal 20-30 amino acids of various GYF domains, supporting the idea that this part of the domain is structurally but not functionally important.

In molecular biology, the H2TH domain is a DNA-binding domain found in DNA glycosylase/AP lyase enzymes, which are involved in base excision repair of DNA damaged by oxidation or by mutagenic agents. Most damage to bases in DNA is repaired by the base excision repair pathway. These enzymes are primarily from bacteria, and have both DNA glycosylase activity EC 3.2.2.- and AP lyase activity EC 4.2.99.18. Examples include formamidopyrimidine-DNA glycosylases and endonuclease VIII (Nei).

DEPDC5 is a human protein of poorly understood function but has been associated with cancer in several studies. It is encoded by a gene of the same name, located on chromosome 22.

In molecular biology, the Tymovirus coat protein refers to the protein coat of a virus order, named Tymovirales. More specifically this protein signature is found only in coat proteins from the related tymoviruses. The coat protein (CP) is also known as the virion protein. The virus coat is composed of 180 copies of the coat protein arranged in an icosahedral shell.

Solenoid protein domains are a highly modular type of protein domain. They consist of a chain of nearly identical folds, often simply called tandem repeats. They are extremely common among all types of proteins, though exact figures are unknown.




















