
Genome projects are scientific endeavours that ultimately aim to determine the complete genome sequence of an organism and to annotate protein-coding genes and other important genome-encoded features. The genome sequence of an organism includes the collective DNA sequences of each chromosome in the organism. For a bacterium containing a single chromosome, a genome project will aim to map the sequence of that chromosome. For the human species, whose genome includes 22 pairs of autosomes and 2 sex chromosomes, a complete genome sequence will involve 46 separate chromosome sequences.
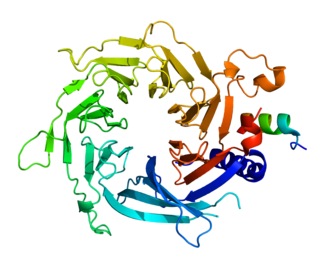
Histone-binding protein RBBP7 is a protein that in humans is encoded by the RBBP7 gene.

NAD-dependent deacetylase sirtuin 2 is an enzyme that in humans is encoded by the SIRT2 gene. SIRT2 is an NAD+ -dependent deacetylase. Studies of this protein have often been divergent, highlighting the dependence of pleiotropic effects of SIRT2 on cellular context. The natural polyphenol resveratrol is known to exert opposite actions on neural cells according to their normal or cancerous status. Similar to other sirtuin family members, SIRT2 displays a ubiquitous distribution. SIRT2 is expressed in a wide range of tissues and organs and has been detected particularly in metabolically relevant tissues, including the brain, muscle, liver, testes, pancreas, kidney, and adipose tissue of mice. Of note, SIRT2 expression is much higher in the brain than all other organs studied, particularly in the cortex, striatum, hippocampus, and spinal cord.

Histone-lysine N-methyltransferase SETDB1 is an enzyme that in humans is encoded by the SETDB1 gene. SETDB1 is also known as KMT1E or H3K9 methyltransferase ESET.

Centromere protein J is a protein that in humans is encoded by the CENPJ gene. It is also known as centrosomal P4.1-associated protein (CPAP). During cell division, this protein plays a structural role in the maintenance of centrosome integrity and normal spindle morphology, and it is involved in microtubule disassembly at the centrosome. This protein can function as a transcriptional coactivator in the Stat5 signaling pathway and also as a coactivator of NF-kappaB-mediated transcription, likely via its interaction with the coactivator p300/CREB-binding protein.

Syntaxin-8 is a protein that in humans is encoded by the STX8 gene. Syntaxin 8 directly interacts with HECTd3 and has similar subcellular localization. The protein has been shown to form the SNARE complex with syntaxin 7, vti1b and endobrevin. These function as the machinery for the homotypic fusion of late endosomes.

Monocarboxylate transporter 8 (MCT8) is an active transporter protein that in humans is encoded by the SLC16A2 gene.

Ubiquitin-associated protein 1 is a protein that in humans is encoded by the UBAP1 gene.

AT-rich interactive domain-containing protein 2 (ARID2) is a protein that in humans is encoded by the ARID2 gene.

Shugoshin 1 or Shugoshin-like 1, is a protein that in humans is encoded by the SGO1 gene.

Kaptin is a protein that in humans is encoded by the KPTN gene.

Lysine-specific demethylase 4C is an enzyme that in humans is encoded by the KDM4C gene.

Condensin complex subunit 2 also known as chromosome-associated protein H (CAP-H) or non-SMC condensin I complex subunit H (NCAPH) is a protein that in humans is encoded by the NCAPH gene. CAP-H is a subunit of condensin I, a large protein complex involved in chromosome condensation. Abnormal expression of NCAPH may be linked to various types of carcinogenesis as a prognostic indicator.

TRAF3-interacting JNK-activating modulator is a protein that in humans is encoded by the TRAF3IP3 gene.

Protein transport protein Sec16B also known as regucalcin gene promoter region-related protein p117 (RGPR-p117) and leucine zipper transcription regulator 2 (LZTR2) is a protein that in humans is encoded by the SEC16B gene.

Cytokine receptor-like factor 3 is a protein that in humans is encoded by the CRLF3 gene.

Oxysterol binding protein-like 9 is a protein that in humans is encoded by the OSBPL9 gene.

tripartite motif containing 45, also known as TRIM45, is a human gene.

Transmembrane protein 165 is a protein that in humans is encoded by the TMEM165 gene.
Kang Zhang is a Chinese-American ophthalmologist specializing in ophthalmic genetics and aging processes in the eye. He is currently a Professor of the Faculty of Medicine at Macau University of Science and Technology. He was previously a Professor of Ophthalmology and the Founding Director of the Institute for Genomic Medicine at the University of California, San Diego. Zhang is particularly known for his work on lanosterol, stem cell research, gene editing, and artificial intelligence.

















