Related Research Articles
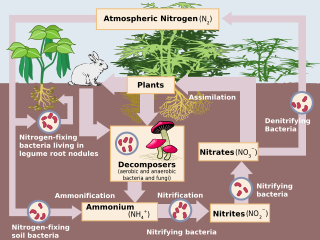
Denitrification is a microbially facilitated process where nitrate (NO3−) is reduced and ultimately produces molecular nitrogen (N2) through a series of intermediate gaseous nitrogen oxide products. Facultative anaerobic bacteria perform denitrification as a type of respiration that reduces oxidized forms of nitrogen in response to the oxidation of an electron donor such as organic matter. The preferred nitrogen electron acceptors in order of most to least thermodynamically favorable include nitrate (NO3−), nitrite (NO2−), nitric oxide (NO), nitrous oxide (N2O) finally resulting in the production of dinitrogen (N2) completing the nitrogen cycle. Denitrifying microbes require a very low oxygen concentration of less than 10%, as well as organic C for energy. Since denitrification can remove NO3−, reducing its leaching to groundwater, it can be strategically used to treat sewage or animal residues of high nitrogen content. Denitrification can leak N2O, which is an ozone-depleting substance and a greenhouse gas that can have a considerable influence on global warming.

Anammox, an abbreviation for "anaerobic ammonium oxidation", is a globally important microbial process of the nitrogen cycle that takes place in many natural environments. The bacteria mediating this process were identified in 1999, and were a great surprise for the scientific community. In the anammox reaction, nitrite and ammonium ions are converted directly into diatomic nitrogen and water.
Denitrifying bacteria are a diverse group of bacteria that encompass many different phyla. This group of bacteria, together with denitrifying fungi and archaea, is capable of performing denitrification as part of the nitrogen cycle. Denitrification is performed by a variety of denitrifying bacteria that are widely distributed in soils and sediments and that use oxidized nitrogen compounds in absence of oxygen as a terminal electron acceptor. They metabolise nitrogenous compounds using various enzymes, turning nitrogen oxides back to nitrogen gas or nitrous oxide.

Sequencing batch reactors (SBR) or sequential batch reactors are a type of activated sludge process for the treatment of wastewater. SBR reactors treat wastewater such as sewage or output from anaerobic digesters or mechanical biological treatment facilities in batches. Oxygen is bubbled through the mixture of wastewater and activated sludge to reduce the organic matter. The treated effluent may be suitable for discharge to surface waters or possibly for use on land.
Nitrifying bacteria are chemolithotrophic organisms that include species of genera such as Nitrosomonas, Nitrosococcus, Nitrobacter, Nitrospina, Nitrospira and Nitrococcus. These bacteria get their energy from the oxidation of inorganic nitrogen compounds. Types include ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria (NOB). Many species of nitrifying bacteria have complex internal membrane systems that are the location for key enzymes in nitrification: ammonia monooxygenase, hydroxylamine oxidoreductase, and nitrite oxidoreductase.
Paracoccus denitrificans, is a coccoid bacterium known for its nitrate reducing properties, its ability to replicate under conditions of hypergravity and for being a relative of the eukaryotic mitochondrion.
Nitrospira translate into “a nitrate spiral” is a genus of bacteria within the monophyletic clade of the Nitrospirota phylum. The first member of this genus was described 1986 by Watson et al. isolated from the Gulf of Maine. The bacterium was named Nitrospira marina. Populations were initially thought to be limited to marine ecosystems, but it was later discovered to be well-suited for numerous habitats, including activated sludge of wastewater treatment systems, natural biological marine settings, water circulation biofilters in aquarium tanks, terrestrial systems, fresh and salt water ecosystems, and hot springs. Nitrospira is a ubiquitous bacterium that plays a role in the nitrogen cycle by performing nitrite oxidation in the second step of nitrification. Nitrospira live in a wide array of environments including but not limited to, drinking water systems, waste treatment plants, rice paddies, forest soils, geothermal springs, and sponge tissue. Despite being abundant in many natural and engineered ecosystems Nitrospira are difficult to culture, so most knowledge of them is from molecular and genomic data. However, due to their difficulty to be cultivated in laboratory settings, the entire genome was only sequenced in one species, Nitrospira defluvii. In addition, Nitrospira bacteria's 16s rRNA sequences are too dissimilar to use for PCR primers, thus some members go unnoticed. In addition, members of Nitrospira with the capabilities to perform complete nitrification has also been discovered and cultivated.

SHARON is a sewage treatment process. A partial nitrification process of sewage treatment used for the removal of ammonia and organic nitrogen components from wastewater flow streams. The process results in stable nitrite formation, rather than complete oxidation to nitrate. Nitrate formation by nitrite oxidising bacteria (NOB) is prevented by adjusting temperature, pH, and retention time to select for nitrifying ammonia oxidising bacteria (AOB). Denitrification of waste streams utilizing SHARON reactors can proceed with an anoxic reduction, such as anammox.
Candidatus Accumulibacter phosphatis (CAP) is an unclassified type of Betaproteobacteria that is a common bacterial community member of sewage treatment and wastewater treatment plants performing enhanced biological phosphorus removal (EBPR) and is a polyphosphate-accumulating organism. The role of CAP in EBPR was elucidated using culture-independent approaches such as 16S rRNA clone banks that showed the Betaproteobacteria dominated lab-scale EBPR reactors. Further work using clone banks and fluorescence in situ hybridization identified a group of bacteria, closely related to Rhodocyclus as the dominant member of lab-scale communities.
"Candidatus Scalindua" is a bacterial genus, and a proposed member of the order Planctomycetales. These bacteria lack peptidoglycan in their cell wall and have a compartmentalized cytoplasm. They are ammonium oxidizing bacteria found in marine environments.
Candidatus Brocadia fulgida is a bacterial species that performs the anammox process. Fatty acids constitute an enrichment culture for B. fulgida. The species' 16S ribosomal RNA sequence has been determined. During the anammox process, it oxidizes acetate at the highest rate and outcompetes other anammox bacteria, which indicates that it does not incorporate acetate directly into its biomass like other anammox bacteria.
Comammox is the name attributed to an organism that can convert ammonia into nitrite and then into nitrate through the process of nitrification. Nitrification has traditionally thought to be a two-step process, where ammonia-oxidizing bacteria and archaea oxidize ammonia to nitrite and then nitrite-oxidizing bacteria convert to nitrate. Complete conversion of ammonia into nitrate by a single microorganism was first predicted in 2006. In 2015 the presence of microorganisms that could carry out both conversion processes was discovered within the genus Nitrospira, and the nitrogen cycle was updated. Within the genus Nitrospira, the major ecosystems comammox are primarily found in natural aquifers and engineered ecosystems.
Thioalkalivibrio is a Gram-negative, mostly halophilic bacterial genus of the family Ectothiorhodospiraceae.
Dissimilatory nitrate reduction to ammonium (DNRA), also known as nitrate/nitrite ammonification, is the result of anaerobic respiration by chemoorganoheterotrophic microbes using nitrate (NO3−) as an electron acceptor for respiration. In anaerobic conditions microbes which undertake DNRA oxidise organic matter and use nitrate (rather than oxygen) as an electron acceptor, reducing it to nitrite, then ammonium (NO3−→NO2−→NH4+).
An oxygen minimum zone (OMZ) is characterized as an oxygen-deficient layer in the world's oceans. Typically found between 200m to 1500m deep below regions of high productivity, such as the western coasts of continents. OMZs can be seasonal following the spring-summer upwelling season. Upwelling of nutrient-rich water leads to high productivity and labile organic matter, that is respired by heterotrophs as it sinks down the water column. High respiration rates deplete the oxygen in the water column to concentrations of 2 mg/L or less forming the OMZ. OMZs are expanding, with increasing ocean deoxygenation. Under these oxygen-starved conditions, energy is diverted from higher trophic levels to microbial communities that have evolved to use other biogeochemical species instead of oxygen, these species include Nitrate, Nitrite, Sulphate etc. Several Bacteria and Archea have adapted to live in these environments by using these alternate chemical species and thrive. The most abundant phyla in OMZs are Pseudomonadota, Bacteroidota, Actinomycetota, and Planctomycetota.

NC10 is a bacterial phylum with candidate status, meaning its members remain uncultured to date. The difficulty in producing lab cultures may be linked to low growth rates and other limiting growth factors.
CandidatusAnammoxoglobus propionicus is an anammox bacteria that is taxonomically in the phylum of Planctomycetota. Anammoxoglobus propionicus is an interest to many researchers due to its ability to reduce nitrite and oxidize ammonium into nitrogen gas and water.

Candidatus "Methylomirabilis oxyfera" is a candidate species of Gram-negative bacteria belonging to the NC10 phylum, characterized for its capacity to couple anaerobic methane oxidation with nitrite reduction in anoxic environments. To acquire oxygen for methane oxidation, M. oxyfera utilizes an intra-aerobic pathway through the reduction of nitrite (NO2) to dinitrogen (N2) and oxygen.
"Candidatus Brocadia" is a candidatus genus of bacteria, meaning that while it is well-characterized, it has not been grown as a pure culture yet. Due to this, much of what is known about Candidatus species has been discovered using culture-independent techniques such as metagenomic sequence analysis.
Anammox is a wastewater treatment technique that removes nitrogen using anaerobic ammonium oxidation (anammox). This process is performed by anammox bacteria which are autotrophic, meaning they do not need organic carbon for their metabolism to function. Instead, the metabolism of anammox bacteria convert ammonium and nitrite into dinitrogen gas. Anammox bacteria are a wastewater treatment technique and wastewater treatment facilities are in the process of implementing anammox-based technologies to further enhance ammonia and nitrogen removal.
References
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 Schmid, Markus; Walsh, Kerry; Webb, Rick; Rijpstra, W. Irene; van de Pas-Schoonen, Katinka; Verbruggen, Mark Jan; Hill, Thomas; Moffett, Bruce; Fuerst, John; Schouten, Stefan; Sinninghe Damsté, Jaap S.; Harris, James; Shaw, Phil; Jetten, Mike; Strous, Marc (January 2003). "Candidatus "Scalindua brodae", sp. nov., Candidatus "Scalindua wagneri", sp. nov., Two New Species of Anaerobic Ammonium Oxidizing Bacteria". Systematic and Applied Microbiology. 26 (4): 529–538. doi:10.1078/072320203770865837. PMID 14666981.
- 1 2 3 4 5 6 Jetten, Mike; Schmid, Markus; Schmidt, Ingo; van Loosdrescht, Mark; Abma, Wiebe; Kuenen, J. Gijs; Mulder, Jan-Willem; Strous, Marc (2004). Biodiversity and application of anaerobic ammonium-oxidizing bacteria. pp. 21–26. ISBN 9789058096531 . Retrieved 15 February 2016.
{{cite book}}:|journal=ignored (help) - 1 2 van Niftrik, Laura A.; Fuerst, John A.; Damste, Jaap S. Sinninghe; Kuenen, J. Gijs; Jetten, Mike S.M.; Strous, Marc (April 2004). "The anammoxosome: an intracytoplasmic compartment in anammox bacteria". FEMS Microbiology Letters. 233 (1): 7–13. doi: 10.1016/j.femsle.2004.01.044 . PMID 15098544.
- 1 2 3 4 5 6 Nakajima, J; Sakka, M; Kimura, T; Furukawa, K; Sakka, K (2008). "Enrichment of anammox bacteria from marine environment for the construction of a bioremediation reactor". Applied Microbiology and Biotechnology. 77 (5): 1159–1166. doi:10.1007/s00253-007-1247-7. PMID 17965857. S2CID 20103740.
- 1 2 "Scalindua wagneri". NCBI. Retrieved 10 March 2015.
- ↑ van de Vossenberg, Jack; Woebken, Dagmar; Maalcke, Wouter J.; Wessels, Hans J. C. T.; Dutilh, Bas E.; Kartal, Boran; Janssen-Megens, Eva M.; Roeselers, Guus; Yan, Jia; Speth, Daan; Gloerich, Jolein; Geerts, Wim; van der Biezen, Erwin; Pluk, Wendy; Francoijs, Kees-Jan; Russ, Lina; Lam, Phyllis; Malfatti, Stefanie A.; Tringe, Susannah Green; Haaijer, Suzanne C. M.; Op den Camp, Huub J. M.; Stunnenberg, Henk G.; Amann, Rudi; Kuypers, Marcel M. M.; Jetten, Mike S. M. (May 2013). "The metagenome of the marine anammox bacterium 'Scalindua profunda' illustrates the versatility of this globally important nitrogen cycle bacterium". Environmental Microbiology. 15 (5): 1275–1289. doi:10.1111/j.1462-2920.2012.02774.x. PMC 3655542 . PMID 22568606.
- 1 2 Li, Hui; Chen, Shuo; Mu, Bo-Zhong; Gu, Ji-Dong (26 August 2010). "Molecular Detection of Anaerobic Ammonium-Oxidizing (Anammox) Bacteria in High-Temperature Petroleum Reservoirs". Microbial Ecology. 60 (4): 771–783. doi:10.1007/s00248-010-9733-3. PMC 2974184 . PMID 20740282.
- ↑ Penton, C. R.; Devol, A. H.; Tiedje, J. M. (4 October 2006). "Molecular Evidence for the Broad Distribution of Anaerobic Ammonium-Oxidizing Bacteria in Freshwater and Marine Sediments". Applied and Environmental Microbiology. 72 (10): 6829–6832. Bibcode:2006ApEnM..72.6829P. doi:10.1128/AEM.01254-06. PMC 1610322 . PMID 17021238.
- 1 2 Hu, Z.; Lotti, T.; de Kreuk, M.; Kleerebezem, R.; van Loosdrecht, M.; Kruit, J.; Jetten, M. S. M.; Kartal, B. (15 February 2013). "Nitrogen Removal by a Nitritation-Anammox Bioreactor at Low Temperature". Applied and Environmental Microbiology. 79 (8): 2807–2812. Bibcode:2013ApEnM..79.2807H. doi:10.1128/AEM.03987-12. PMC 3623191 . PMID 23417008.
- ↑ Wright, Jody J.; Konwar, Kishori M.; Hallam, Steven J. (14 May 2012). "Microbial ecology of expanding oxygen minimum zones". Nature Reviews Microbiology. 10 (6): 381–394. doi:10.1038/nrmicro2778. PMID 22580367. S2CID 1467645.
- ↑ Moore, C. M.; Mills, M. M.; Arrigo, K. R.; Berman-Frank, I.; Bopp, L.; Boyd, P. W.; Galbraith, E. D.; Geider, R. J.; Guieu, C.; Jaccard, S. L.; Jickells, T. D.; La Roche, J.; Lenton, T. M.; Mahowald, N. M.; Marañón, E.; Marinov, I.; Moore, J. K.; Nakatsuka, T.; Oschlies, A.; Saito, M. A.; Thingstad, T. F.; Tsuda, A.; Ulloa, O. (31 March 2013). "Processes and patterns of oceanic nutrient limitation". Nature Geoscience. 6 (9): 701–710. Bibcode:2013NatGe...6..701M. CiteSeerX 10.1.1.397.5625 . doi:10.1038/ngeo1765.