Related Research Articles

Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions,either by performing the function itself or by forming a template for the production of proteins. RNA and deoxyribonucleic acid (DNA) are nucleic acids. The nucleic acids constitute one of the four major macromolecules essential for all known forms of life. RNA is assembled as a chain of nucleotides. Cellular organisms use messenger RNA (mRNA) to convey genetic information that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome.

The RNA world is a hypothetical stage in the evolutionary history of life on Earth in which self-replicating RNA molecules proliferated before the evolution of DNA and proteins. The term also refers to the hypothesis that posits the existence of this stage. Alexander Rich first proposed the concept of the RNA world in 1962,and Walter Gilbert coined the term in 1986.
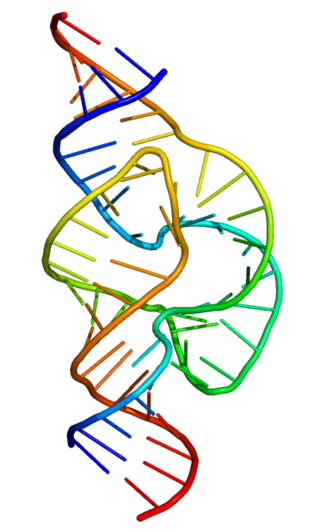
Ribozymes are RNA molecules that have the ability to catalyze specific biochemical reactions,including RNA splicing in gene expression,similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material and a biological catalyst,and contributed to the RNA world hypothesis,which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems.

In molecular biology,a riboswitch is a regulatory segment of a messenger RNA molecule that binds a small molecule,resulting in a change in production of the proteins encoded by the mRNA. Thus,an mRNA that contains a riboswitch is directly involved in regulating its own activity,in response to the concentrations of its effector molecule. The discovery that modern organisms use RNA to bind small molecules,and discriminate against closely related analogs,expanded the known natural capabilities of RNA beyond its ability to code for proteins,catalyze reactions,or to bind other RNA or protein macromolecules.
The peptidyl transferase center is an aminoacyltransferase ribozyme located in the large subunit of the ribosome. It forms peptide bonds between adjacent amino acids during the translation process of protein biosynthesis. It is also responsible for peptidyl-tRNA hydrolysis,allowing the release of the synthesized peptide chain at the end of translation. Peptidyl transferase activity is not mediated by any ribosomal proteins,but entirely by ribosomal RNA (rRNA). The peptidyl transferase center is a significant piece of evidence supporting the RNA World hypothesis.
The history of molecular biology begins in the 1930s with the convergence of various,previously distinct biological and physical disciplines:biochemistry,genetics,microbiology,virology and physics. With the hope of understanding life at its most fundamental level,numerous physicists and chemists also took an interest in what would become molecular biology.

Joan Elaine Argetsinger Steitz is Sterling Professor of Molecular Biophysics and Biochemistry at Yale University and Investigator at the Howard Hughes Medical Institute. She is known for her discoveries involving RNA,including ground-breaking insights into how ribosomes interact with messenger RNA by complementary base pairing and that introns are spliced by small nuclear ribonucleic proteins (snRNPs),which occur in eukaryotes. In September 2018,Steitz won the Lasker-Koshland Award for Special Achievement in Medical Science. The Lasker award is often referred to as the 'American Nobel' because 87 of the former recipients have gone on to win Nobel prizes.
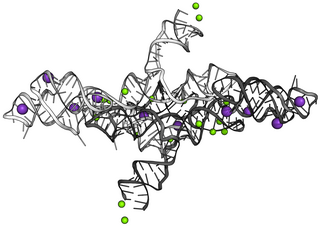
The Varkud satellite (VS) ribozyme is an RNA enzyme that carries out the cleavage of a phosphodiester bond.

The glucosamine-6-phosphate riboswitch ribozyme is an RNA structure that resides in the 5' untranslated region (UTR) of the mRNA transcript of the glmS gene. This RNA regulates the glmS gene by responding to concentrations of a specific metabolite,glucosamine-6-phosphate (GlcN6P),in addition to catalyzing a self-cleaving chemical reaction upon activation. This cleavage leads to the degradation of the mRNA that contains the ribozyme,and lowers production of GlcN6P. The glmS gene encodes for an enzyme glutamine-fructose-6-phosphate amidotransferase,which catalyzes the formation of GlcN6P,a compound essential for cell wall biosynthesis,from fructose-6-phosphate and glutamine. Thus,when GlcN6P levels are high,the glmS ribozyme is activated and the mRNA transcript is degraded but in the absence of GlcN6P the gene continues to be translated into glutamine-fructose-6-phosphate amidotransferase and GlcN6P is produced. GlcN6P is a cofactor for this cleavage reaction,as it directly participates as an acid-base catalyst. This RNA is the first riboswitch also found to be a self-cleaving ribozyme and,like many others,was discovered using a bioinformatics approach.

The bacterial glycine riboswitch is an RNA element that can bind the amino acid glycine. Glycine riboswitches usually consist of two metabolite-binding aptamer domains with similar structures in tandem. The aptamers were originally thought to cooperatively bind glycine to regulate the expression of downstream genes. In Bacillus subtilis,this riboswitch is found upstream of the gcvT operon which controls glycine degradation. It is thought that when glycine is in excess it will bind to both aptamers to activate these genes and facilitate glycine degradation.
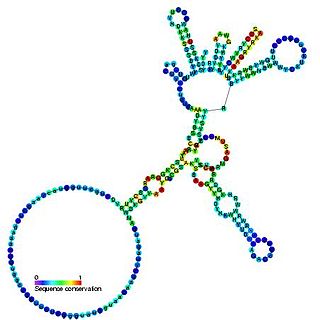
Group I introns are large self-splicing ribozymes. They catalyze their own excision from mRNA,tRNA and rRNA precursors in a wide range of organisms. The core secondary structure consists of nine paired regions (P1-P9). These fold to essentially two domains –the P4-P6 domain and the P3-P9 domain. The secondary structure mark-up for this family represents only this conserved core. Group I introns often have long open reading frames inserted in loop regions.

50S is the larger subunit of the 70S ribosome of prokaryotes,i.e. bacteria and archaea. It is the site of inhibition for antibiotics such as macrolides,chloramphenicol,clindamycin,and the pleuromutilins. It includes the 5S ribosomal RNA and 23S ribosomal RNA.
The Lariat capping ribozyme is a ~180 nt ribozyme with an apparent resemblance to a group I ribozyme. It is found within a complex type of group I introns also termed twin-ribozyme introns. Rather than splicing,it catalyses a branching reaction in which the 2'OH of an internal residue is involved in a nucleophilic attack at a nearby phosphodiester bond. As a result,the RNA is cleaved at an internal processing site (IPS),leaving a 3'OH and a downstream product with a 3 nt lariat at its 5' end. The lariat has the first and the third nucleotide joined by a 2',5' phosphodiester bond and is referred to as 'the lariat cap' because it caps an intron-encoded mRNA. The resulting lariat cap seems to contribute by increasing the half-life of the HE mRNA,thus conferring an evolutionary advantage to the HE.

Thomas Arthur Steitz was an American biochemist,a Sterling Professor of Molecular Biophysics and Biochemistry at Yale University,and investigator at the Howard Hughes Medical Institute,best known for his pioneering work on the ribosome.
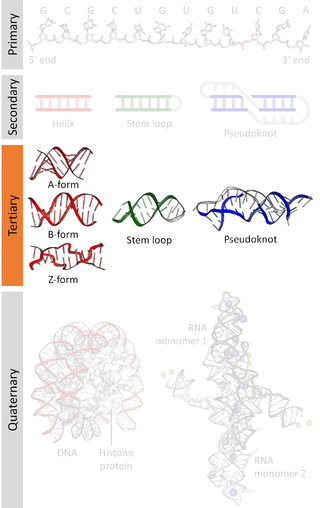
Nucleic acid tertiary structure is the three-dimensional shape of a nucleic acid polymer. RNA and DNA molecules are capable of diverse functions ranging from molecular recognition to catalysis. Such functions require a precise three-dimensional structure. While such structures are diverse and seemingly complex,they are composed of recurring,easily recognizable tertiary structural motifs that serve as molecular building blocks. Some of the most common motifs for RNA and DNA tertiary structure are described below,but this information is based on a limited number of solved structures. Many more tertiary structural motifs will be revealed as new RNA and DNA molecules are structurally characterized.

Cyclic di-GMP-II riboswitches form a class of riboswitches that specifically bind cyclic di-GMP,a second messenger used in multiple bacterial processes such as virulence,motility and biofilm formation. Cyclic di-GMP II riboswitches are structurally unrelated to cyclic di-GMP-I riboswitches,though they have the same function.
Numerous key discoveries in biology have emerged from studies of RNA,including seminal work in the fields of biochemistry,genetics,microbiology,molecular biology,molecular evolution,and structural biology. As of 2010,30 scientists have been awarded Nobel Prizes for experimental work that includes studies of RNA. Specific discoveries of high biological significance are discussed in this article.
Taekjip Ha is a South Korean-born American biophysicist who is currently a Bloomberg Distinguished Professor of Biophysics and Biomedical Engineering at Johns Hopkins University. He was previously the Gutgsell Professor of Physics,at University of Illinois at Urbana-Champaign where he was the principal investigator of Single Molecule Nanometry group. He is also a Howard Hughes Medical Institute investigator.
Paul B. Sigler was the Henry Ford II Professor of Molecular Biophysics and Biochemistry at Yale University. Major awards included membership in the National Academy of Sciences,HHMI Investigator status,and Guggenheim and Helen Hay Whitney Fellowships. He is noted for pioneering studies of Phospholipase A2 and trp repressor amongst many others.
Susan A. Martinis is an American biochemist. She has co-authored over 57 publications in peer reviewed journals and scientific book chapters. Her expertise is in protein:RNA interactions and aminoacyl tRNA synthetases. As of 2019,she is the Vice Chancellor for Research and Innovation at the University of Illinois at Urbana-Champaign.
References
- ↑ "Site-specific cleavage of genomic DNA mediated by triple helix formation - ProQuest". www.proquest.com. Retrieved 2024-12-06.
- ↑ Peart, Karen (6 November 2019). "Scott Strobel named Yale provost". YaleNews.
- 1 2 "2018 Meritorious Service Award – Scott A. Strobel". YSEA. 2021-07-19. Retrieved 2024-01-12.
- 1 2 "Scott Strobel, PhD". medicine.yale.edu. Retrieved 2024-01-12.
- ↑ Strobel SA; Shetty K. (1997). "Defining the chemical groups essential for Tetrahymena group I intron function by nucleotide analog interference mapping". Proceedings of the National Academy of Sciences USA. 94 (7): 2903–2908. Bibcode:1997PNAS...94.2903S. doi: 10.1073/pnas.94.7.2903 . PMC 20295 . PMID 9096319.
- ↑ "Yale Scientists Visualize Molecular Detail Of RNA Splicing Complex" (June 3, 2004). ScienceDaily Retrieved October 16, 2012
- ↑ Adams PL; Stahley MR; Kosek AB; Wang J; Strobel SA (2004). "Crystal Structure of a Self-Splicing Group I Intron with Both Exons". Nature. 430 (6995): 45–50. Bibcode:2004Natur.430...45A. doi:10.1038/nature02642. PMID 15175762. S2CID 4387064.
- ↑ Cochrane JC; Lipchock SV; Smith KD; Strobel SA (2009). "Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme". Biochemistry. 48 (15): 3239–3246. doi:10.1021/bi802069p. PMC 2854835 . PMID 19228039.
- ↑ Smith KD; Lipchock SV; Ames TD; Wang J; Breaker RR; Strobel SA (2009). "Structural basis of ligand binding by a c-di-GMP riboswitch". Nature Structural & Molecular Biology. 16 (12): 1218–1223. doi:10.1038/nsmb.1702. PMC 2850612 . PMID 19898477.
- ↑ Schmeing TM; Huang KS; Strobel SA; Steitz TA (2005). "An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA". Nature. 438 (7067): 520–524. Bibcode:2005Natur.438..520M. doi:10.1038/nature04152. PMID 16306996. S2CID 4333559.
- ↑ Russell, JR; Huang, J; Anand, P; Kucera, K; Sandoval, AG; Dantzler, KW; Hickman, D; Jee, J; Kimovec, FM; Koppstein, D; Marks, DH; Mittermiller, PA; Núñez, SJ; Santiago, M; Townes, MA; Vishnevetsky, M; Williams, NE; Vargas, MP; Boulanger, LA; Bascom-Slack, C; Strobel, SA (September 2011). "Biodegradation of polyester polyurethane by endophytic fungi". Applied and Environmental Microbiology. 77 (17): 6076–6084. doi:10.1128/AEM.00521-11. PMC 3165411 . PMID 21764951.
- ↑ Arnaud, Celia (10 November 2008). "Into The Woods". C&EN. Retrieved 24 September 2012.
- ↑ Gasso, Jordi (11 October 2010). "Prof. branches out with Yale Bowls". Yale Daily News. Retrieved 30 November 2015.