Related Research Articles
Protein targeting or protein sorting is the biological mechanism by which proteins are transported to their appropriate destinations within or outside the cell. Proteins can be targeted to the inner space of an organelle, different intracellular membranes, the plasma membrane, or to the exterior of the cell via secretion. Information contained in the protein itself directs this delivery process. Correct sorting is crucial for the cell; errors or dysfunction in sorting have been linked to multiple diseases.
The translocon is a complex of proteins associated with the translocation of polypeptides across membranes. In eukaryotes the term translocon most commonly refers to the complex that transports nascent polypeptides with a targeting signal sequence into the interior space of the endoplasmic reticulum (ER) from the cytosol. This translocation process requires the protein to cross a hydrophobic lipid bilayer. The same complex is also used to integrate nascent proteins into the membrane itself. In prokaryotes, a similar protein complex transports polypeptides across the (inner) plasma membrane or integrates membrane proteins. In either case, the protein complex are formed from Sec proteins, with the heterotrimeric Sec61 being the channel. In prokaryotes, the homologous channel complex is known as SecYEG.
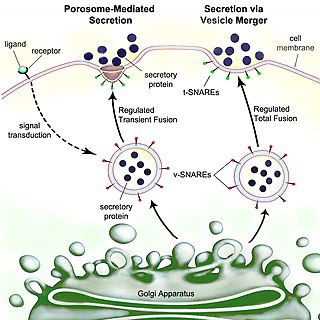
Secretion is the movement of material from one point to another, such as a secreted chemical substance from a cell or gland. In contrast, excretion is the removal of certain substances or waste products from a cell or organism. The classical mechanism of cell secretion is via secretory portals at the plasma membrane called porosomes. Porosomes are permanent cup-shaped lipoprotein structures embedded in the cell membrane, where secretory vesicles transiently dock and fuse to release intra-vesicular contents from the cell.
Sec61, termed SecYEG in prokaryotes, is a membrane protein complex found in all domains of life. As the core component of the translocon, it transports proteins to the endoplasmic reticulum in eukaryotes and out of the cell in prokaryotes. It is a doughnut-shaped pore through the membrane with 3 different subunits (heterotrimeric), SecY (α), SecE (γ), and SecG (β). It has a region called the plug that blocks transport into or out of the ER. This plug is displaced when the hydrophobic region of a nascent polypeptide interacts with another region of Sec61 called the seam, allowing translocation of the polypeptide into the ER lumen.
The twin-arginine translocation pathway is a protein export, or secretion pathway found in plants, bacteria, and archaea. In contrast to the Sec pathway which transports proteins in an unfolded manner, the Tat pathway serves to actively translocate folded proteins across a lipid membrane bilayer. In plants, the Tat translocase is located in the thylakoid membrane of the chloroplast, where it acts to export proteins into the thylakoid lumen. In bacteria, the Tat translocase is found in the cytoplasmic membrane and serves to export proteins to the cell envelope, or to the extracellular space. The existence of a Tat translocase in plant mitochondria is also proposed.

Mitochondrial membrane transport proteins, also known as mitochondrial carrier proteins, are proteins which exist in the membranes of mitochondria. They serve to transport molecules and other factors, such as ions, into or out of the organelles. Mitochondria contain both an inner and outer membrane, separated by the inter-membrane space, or inner boundary membrane. The outer membrane is porous, whereas the inner membrane restricts the movement of all molecules. The two membranes also vary in membrane potential and pH. These factors play a role in the function of mitochondrial membrane transport proteins. There are 53 discovered human mitochondrial membrane transporters, with many others that are known to still need discovered.
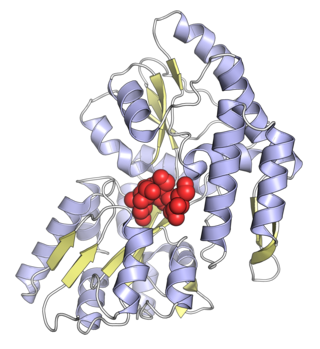
Maltose-binding protein (MBP) is a part of the maltose/maltodextrin system of Escherichia coli, which is responsible for the uptake and efficient catabolism of maltodextrins. It is a complex regulatory and transport system involving many proteins and protein complexes. MBP has an approximate molecular mass of 42.5 kilodaltons.
Translocase is a general term for a protein that assists in moving another molecule, usually across a cell membrane. These enzymes catalyze the movement of ions or molecules across membranes or their separation within membranes. The reaction is designated as a transfer from “side 1” to “side 2” because the designations “in” and “out”, which had previously been used, can be ambiguous. Translocases are the most common secretion system in Gram positive bacteria.
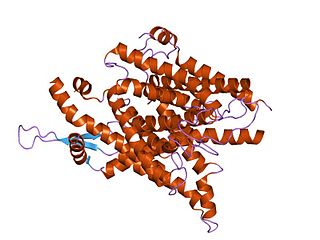
The SecY protein is the main transmembrane subunit of the bacterial Sec export pathway and of a protein-secreting ATPase complex, also known as a SecYEG translocon. Homologs of the SecYEG complex are found in eukaryotes and in archaea, where the subunit is known as Sec61α.
The translocase of the inner membrane (TIM) is a complex of proteins found in the inner membrane of the mitochondrion. Components of the TIM complex facilitate the translocation of proteins across the inner membrane and into the mitochondrial matrix. They also facilitate the insertion of proteins into the inner mitochondrial membrane, where they must reside in order to function, these mainly include members of the mitochondrial carrier family of proteins.
The SecA protein is a cell membrane associated subunit of the bacterial Sec or Type II secretory pathway, a system which is responsible for the secretion of proteins through the cell membrane. Within this system the SecA ATPase forms a translocase complex with the SecYEG channel, thereby driving the movement of the protein substrate across the membrane.
Chaperone-usher fimbriae (CU) are linear, unbranching, outer-membrane pili secreted by gram-negative bacteria through the chaperone-usher system rather than through type IV secretion or extracellular nucleation systems. These fimbriae are built up out of modular pilus subunits, which are transported into the periplasm in a Sec dependent manner. Chaperone-usher secreted fimbriae are important pathogenicity factors facilitating host colonisation, localisation and biofilm formation in clinically important species such as uropathogenic Escherichia coli and Pseudomonas aeruginosa.
Gabriel Waksman FMedSci, FRS, is Courtauld professor of biochemistry and molecular biology at University College London (UCL), and professor of structural and molecular biology at Birkbeck College, University of London. He is the director of the Institute of Structural and Molecular Biology (ISMB) at UCL and Birkbeck, head of the Department of Structural and Molecular Biology at UCL, and head of the Department of Biological Sciences at Birkbeck.
The type 2 secretion system is a type of protein secretion machinery found in various species of Gram-negative bacteria, including many human pathogens such as Pseudomonas aeruginosa and Vibrio cholerae. The type II secretion system is one of six protein secretory systems commonly found in Gram-negative bacteria, along with the type I, type III, and type IV secretion systems, as well as the chaperone/usher pathway, the autotransporter pathway/type V secretion system, and the type VI secretion system. Like these other systems, the type II secretion system enables the transport of cytoplasmic proteins across the lipid bilayers that make up the cell membranes of Gram-negative bacteria. Secretion of proteins and effector molecules out of the cell plays a critical role in signaling other cells and in the invasion and parasitism of host cells.
The Monovalent Cation (K+ or Na+):Proton Antiporter-3 (CPA3) Family (TC# 2.A.63) is a member of the Na+ transporting Mrp superfamily. The CPA3 family consists of bacterial multicomponent K+:H+ and Na+:H+ antiporters. The best characterized systems are the PhaABCDEFG system of Sinorhizobium meliloti (TC# 2.A.63.1.1) that functions in pH adaptation and as a K+ efflux system, and the MnhABCDEFG system of Staphylococcus aureus (TC# 2.A.63.1.3) that functions as a Na+ efflux Na+:H+ antiporter.

Bacterial secretion systems are protein complexes present on the cell membranes of bacteria for secretion of substances. Specifically, they are the cellular devices used by pathogenic bacteria to secrete their virulence factors to invade the host cells. They can be classified into different types based on their specific structure, composition and activity. Generally, proteins can be secreted through two different processes. One process is a one-step mechanism in which proteins from the cytoplasm of bacteria are transported and delivered directly through the cell membrane into the host cell. Another involves a two-step activity in which the proteins are first transported out of the inner cell membrane, then deposited in the periplasm, and finally through the outer cell membrane into the host cell.
Linda Randall is a Professor Emerita of Biochemistry and Wurdack Chair Emerita of Biological Chemistry at the University of Missouri. Her research has shown unexpected and complex details of the movement of newly made proteins from the cytosol across membranes into the organelles of the cell. In particular, she found that the entire protein was kept unfolded by association with a chaperone and not just directed to cross membranes by its terminal leader sequence. In 1997, she was elected to the National Academy of Sciences of the USA because of the excellence of this work. She has received a number of other honors and awards.
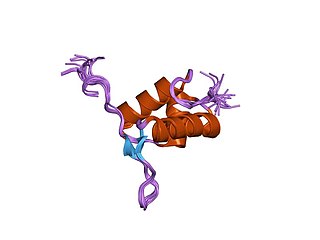
FtsK is a protein in E.Coli involved in bacterial cell division and chromosome segregation. It is one of the largest proteins, consisting of 1329 amino acids. FtsK stands for "Filament temperature sensitive mutant K" because cells expressing a mutant ftsK allele called ftsK44, which encodes an FtsK variant containing an G80A residue change in the second transmembrane segment, fail to divide at high temperatures and form long filaments instead. FtsK, specifically its C-terminal domain, functions as a DNA translocase, interacts with other cell division proteins, and regulates Xer-mediated recombination. FtsK belongs to the AAA superfamily and is present in most bacteria.
Proton-Translocating NAD(P)+ Transhydrogenase (E.C. 7.1.1.1) is an enzyme in that catalyzes the translocation of hydrons that are connected to the redox reaction NADH + NADP+ + H+outside => NAD+ + NADPH + H+inside
Parvulin-like peptidyl-prolyl isomerase (PrsA), also referred to as putative proteinase maturation protein A (PpmA), functions as a molecular chaperone in Gram-positive bacteria, such as B. subtilis, S. aureus, L. monocytogenes and S. pyogenes. PrsA proteins contain a highly conserved parvulin domain that contains peptidyl-prolyl cis-trans isomerase (PPIase) activity capable of catalyzing the bond N-terminal to proline from cis to trans, or vice versa, which is a rate limiting step in protein folding. PrsA homologs also contain a foldase domain suspected to aid in the folding of proteins but, unlike the parvulin domain, is not highly conserved. PrsA proteins are capable of forming multimers in vivo and in vitro and, when dimerized, form a claw-like structure linked by the NC domains. Most Gram-positive bacteria contain only one PrsA-like protein, but some organisms such as L. monocytogenes, B. anthracis and S. pyogenes contain two PrsAs.
References
- 1 2 3 Bolhuis A, Broekhuizen CP, Sorokin A, van Roosmalen ML, Venema G, Bron S, Quax WJ, van Dijl JM (August 1998). "SecDF of Bacillus subtilis, a molecular Siamese twin required for the efficient secretion of proteins". The Journal of Biological Chemistry. 273 (33): 21217–24. doi: 10.1074/jbc.273.33.21217 . PMID 9694879.
- ↑ Arkowitz RA, Wickner W (February 1994). "SecD and SecF are required for the proton electrochemical gradient stimulation of preprotein translocation". The EMBO Journal. 13 (4): 954–63. doi:10.1002/j.1460-2075.1994.tb06340.x. PMC 394897 . PMID 8112309.
- 1 2 Bieker KL, Phillips GJ, Silhavy TJ (June 1990). "The sec and prl genes of Escherichia coli". Journal of Bioenergetics and Biomembranes. 22 (3): 291–310. doi:10.1007/BF00763169. PMID 2202721. S2CID 10694864.
- ↑ Driessen AJ (May 2001). "SecB, a molecular chaperone with two faces" (PDF). Trends in Microbiology. 9 (5): 193–6. doi:10.1016/S0966-842X(01)01980-1. hdl: 11370/4e5ebfac-d58c-42db-b42e-01c22d469a82 . PMID 11336818.
- ↑ Müller JP (July 1999). "Effects of pre-protein overexpression on SecB synthesis in Escherichia coli". FEMS Microbiology Letters. 176 (1): 219–27. doi:10.1111/j.1574-6968.1999.tb13665.x. PMID 10418149.
- ↑ Breyton C, Haase W, Rapoport TA, Kühlbrandt W, Collinson I (August 2002). "Three-dimensional structure of the bacterial protein-translocation complex SecYEG". Nature. 418 (6898): 662–5. Bibcode:2002Natur.418..662B. doi:10.1038/nature00827. PMID 12167867. S2CID 4347664.