
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to express a specific protein in a cell that does not normally express that protein, or to sequence or quantify mRNA molecules using DNA based methods. cDNA that codes for a specific protein can be transferred to a recipient cell for expression, often bacterial or yeast expression systems. cDNA is also generated to analyze transcriptomic profiles in bulk tissue, single cells, or single nuclei in assays such as microarrays, qPCR, and RNA-seq.
Molecular biology is a branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions.

The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993.
Protein engineering is the process of developing useful or valuable proteins through the design and production of unnatural polypeptides, often by altering amino acid sequences found in nature. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles. It has been used to improve the function of many enzymes for industrial catalysis. It is also a product and services market, with an estimated value of $168 billion by 2017.

Reverse transcription polymerase chain reaction (RT-PCR) is a laboratory technique combining reverse transcription of RNA into DNA and amplification of specific DNA targets using polymerase chain reaction (PCR). It is primarily used to measure the amount of a specific RNA. This is achieved by monitoring the amplification reaction using fluorescence, a technique called real-time PCR or quantitative PCR (qPCR). Combined RT-PCR and qPCR are routinely used for analysis of gene expression and quantification of viral RNA in research and clinical settings.
A DNase footprinting assay is a DNA footprinting technique from molecular biology/biochemistry that detects DNA-protein interaction using the fact that a protein bound to DNA will often protect that DNA from enzymatic cleavage. This makes it possible to locate a protein binding site on a particular DNA molecule. The method uses an enzyme, deoxyribonuclease, to cut the radioactively end-labeled DNA, followed by gel electrophoresis to detect the resulting cleavage pattern.
This is a list of topics in molecular biology. See also index of biochemistry articles.

The southwestern blot, is a lab technique that involves identifying as well as characterizing DNA-binding proteins by their ability to bind to specific oligonucleotide probes. Determination of molecular weight of proteins binding to DNA is also made possible by the technique. The name originates from a combination of ideas underlying Southern blotting and Western blotting techniques of which they detect DNA and protein respectively. Similar to other types of blotting, proteins are separated by SDS-PAGE and are subsequently transferred to nitrocellulose membranes. Thereafter southwestern blotting begins to vary with regards to procedure as since the first blotting’s, many more have been proposed and discovered with goals of enhancing results. Former protocols were hampered by the need for large amounts of proteins and their susceptibility to degradation while being isolated.
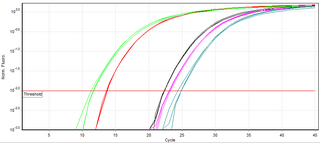
A real-time polymerase chain reaction is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR, not at its end, as in conventional PCR. Real-time PCR can be used quantitatively and semi-quantitatively.

An electrophoretic mobility shift assay (EMSA) or mobility shift electrophoresis, also referred as a gel shift assay, gel mobility shift assay, band shift assay, or gel retardation assay, is a common affinity electrophoresis technique used to study protein–DNA or protein–RNA interactions. This procedure can determine if a protein or mixture of proteins is capable of binding to a given DNA or RNA sequence, and can sometimes indicate if more than one protein molecule is involved in the binding complex. Gel shift assays are often performed in vitro concurrently with DNase footprinting, primer extension, and promoter-probe experiments when studying transcription initiation, DNA gang replication, DNA repair or RNA processing and maturation, as well as pre-mRNA splicing. Although precursors can be found in earlier literature, most current assays are based on methods described by Garner and Revzin and Fried and Crothers.
DNA footprinting is a method of investigating the sequence specificity of DNA-binding proteins in vitro. This technique can be used to study protein-DNA interactions both outside and within cells.

mRNA display is a display technique used for in vitro protein, and/or peptide evolution to create molecules that can bind to a desired target. The process results in translated peptides or proteins that are associated with their mRNA progenitor via a puromycin linkage. The complex then binds to an immobilized target in a selection step. The mRNA-protein fusions that bind well are then reverse transcribed to cDNA and their sequence amplified via a polymerase chain reaction. The result is a nucleotide sequence that encodes a peptide with high affinity for the molecule of interest.
The polymerase chain reaction (PCR) is a commonly used molecular biology tool for amplifying DNA, and various techniques for PCR optimization which have been developed by molecular biologists to improve PCR performance and minimize failure.
ChIP-sequencing, also known as ChIP-seq, is a method used to analyze protein interactions with DNA. ChIP-seq combines chromatin immunoprecipitation (ChIP) with massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins. It can be used to map global binding sites precisely for any protein of interest. Previously, ChIP-on-chip was the most common technique utilized to study these protein–DNA relations.
DNA adenine methyltransferase identification, often abbreviated DamID, is a molecular biology protocol used to map the binding sites of DNA- and chromatin-binding proteins in eukaryotes. DamID identifies binding sites by expressing the proposed DNA-binding protein as a fusion protein with DNA methyltransferase. Binding of the protein of interest to DNA localizes the methyltransferase in the region of the binding site. Adenine methylation does not occur naturally in eukaryotes and therefore adenine methylation in any region can be concluded to have been caused by the fusion protein, implying the region is located near a binding site. DamID is an alternate method to ChIP-on-chip or ChIP-seq.

Promoter bashing is a technique used in molecular biology to identify how certain regions of a DNA strand, commonly promoters, affect the transcription of downstream genes. Under normal circumstances, proteins bind to the promoter and activate or repress transcription. In a promoter bashing assay, specific point mutations or deletions are made in specific regions of the promoter and the transcription of the gene is then measured. The contribution of a region of the promoter can be observed by the level of transcription. If a mutation or deletion changes the level of transcription, then it is known that that region of the promoter may be a binding site or other regulatory element.
The versatility of polymerase chain reaction (PCR) has led to modifications of the basic protocol being used in a large number of variant techniques designed for various purposes. This article summarizes many of the most common variations currently or formerly used in molecular biology laboratories; familiarity with the fundamental premise by which PCR works and corresponding terms and concepts is necessary for understanding these variant techniques.

RIP-chip is a molecular biology technique which combines RNA immunoprecipitation with a microarray. The purpose of this technique is to identify which RNA sequences interact with a particular RNA binding protein of interest in vivo. It can also be used to determine relative levels of gene expression, to identify subsets of RNAs which may be co-regulated, or to identify RNAs that may have related functions. This technique provides insight into the post-transcriptional gene regulation which occurs between RNA and RNA binding proteins.
Epigenomics is the study of the complete set of epigenetic modifications on the genetic material of a cell, known as the epigenome. The field is analogous to genomics and proteomics, which are the study of the genome and proteome of a cell. Epigenetic modifications are reversible modifications on a cell's DNA or histones that affect gene expression without altering the DNA sequence. Epigenomic maintenance is a continuous process and plays an important role in stability of eukaryotic genomes by taking part in crucial biological mechanisms like DNA repair. Plant flavones are said to be inhibiting epigenomic marks that cause cancers. Two of the most characterized epigenetic modifications are DNA methylation and histone modification. Epigenetic modifications play an important role in gene expression and regulation, and are involved in numerous cellular processes such as in differentiation/development and tumorigenesis. The study of epigenetics on a global level has been made possible only recently through the adaptation of genomic high-throughput assays.

The bacterial one-hybrid (B1H) system is a method for identifying the sequence-specific target site of a DNA-binding domain. In this system, a given transcription factor (TF) is expressed as a fusion to a subunit of RNA polymerase. In parallel, a library of randomized oligonucleotides representing potential TF target sequences are cloned into a separate vector containing the selectable genes HIS3 and URA3. If the DNA-binding domain (bait) binds a potential DNA target site (prey) in vivo, it will recruit RNA polymerase to the promoter and activate transcription of the reporter genes in that clone. The two reporter genes, HIS3 and URA3, allow for positive and negative selections, respectively. At the end of the process, positive clones are sequenced and examined with motif-finding tools in order to resolve the favoured DNA target sequence.









