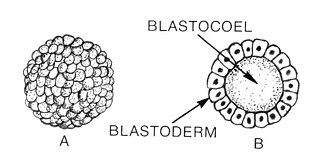
Blastulation is the stage in early animal embryonic development that produces the blastula. The blastula (from Greek βλαστός is a hollow sphere of cells surrounding an inner fluid-filled cavity. Embryonic development begins with a sperm fertilizing an egg cell to become a zygote, which undergoes many cleavages to develop into a ball of cells called a morula. Only when the blastocoel is formed does the early embryo become a blastula. The blastula precedes the formation of the gastrula in which the germ layers of the embryo form.
An oocyte, oöcyte, ovocyte, or rarely ocyte, is a female gametocyte or germ cell involved in reproduction. In other words, it is an immature ovum, or egg cell. An oocyte is produced in the ovary during female gametogenesis. The female germ cells produce a primordial germ cell (PGC), which then undergoes mitosis, forming oogonia. During oogenesis, the oogonia become primary oocytes. An oocyte is a form of genetic material that can be collected for cryoconservation.

Drosophila embryogenesis, the process by which Drosophila embryos form, is a favorite model system for genetics and developmental biology. The study of its embryogenesis unlocked the century-long puzzle of how development was controlled, creating the field of evolutionary developmental biology. The small size, short generation time, and large brood size make it ideal for genetic studies. Transparent embryos facilitate developmental studies. Drosophila melanogaster was introduced into the field of genetic experiments by Thomas Hunt Morgan in 1909.
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature mRNA for translation. In many bacteria, the poly(A) tail promotes degradation of the mRNA. It, therefore, forms part of the larger process of gene expression.
The RNA-induced silencing complex, or RISC, is a multiprotein complex, specifically a ribonucleoprotein, which functions in gene silencing via a variety of pathways at the transcriptional and translational levels. Using single-stranded RNA (ssRNA) fragments, such as microRNA (miRNA), or double-stranded small interfering RNA (siRNA), the complex functions as a key tool in gene regulation. The single strand of RNA acts as a template for RISC to recognize complementary messenger RNA (mRNA) transcript. Once found, one of the proteins in RISC, Argonaute, activates and cleaves the mRNA. This process is called RNA interference (RNAi) and it is found in many eukaryotes; it is a key process in defense against viral infections, as it is triggered by the presence of double-stranded RNA (dsRNA).

Krüppel is a gap gene in Drosophila melanogaster, located on the 2R chromosome, which encodes a zinc finger C2H2 transcription factor. Gap genes work together to establish the anterior-posterior segment patterning of the insect through regulation of the transcription factor encoding pair rule genes. These genes in turn regulate segment polarity genes. Krüppel means "cripple" in German, named for the crippled appearance of mutant larvae, who have failed to develop proper thoracic and anterior segments in the abdominal region. Mutants can also have abdominal mirror duplications.
In developmental biology, midblastula or midblastula transition (MBT) occurs during the blastula stage of embryonic development. During this stage, the embryo is referred to as a blastula. The series of changes to the blastula that characterize the midblastula transition include activation of zygotic gene transcription, slowing of the cell cycle, increased asynchrony in cell division, and an increase in cell motility.
The Let-7 microRNA precursor was identified from a study of developmental timing in C. elegans, and was later shown to be part of a much larger class of non-coding RNAs termed microRNAs. miR-98 microRNA precursor from human is a let-7 family member. Let-7 miRNAs have now been predicted or experimentally confirmed in a wide range of species (MIPF0000002). miRNAs are initially transcribed in long transcripts called primary miRNAs (pri-miRNAs), which are processed in the nucleus by Drosha and Pasha to hairpin structures of about 70 nucleotide. These precursors (pre-miRNAs) are exported to the cytoplasm by exportin5, where they are subsequently processed by the enzyme Dicer to a ~22 nucleotide mature miRNA. The involvement of Dicer in miRNA processing demonstrates a relationship with the phenomenon of RNA interference.

Nanos 3′ UTR translation control element is a cis-regulatory element in the 3′ untranslated region of the messenger RNA which encodes the Nanos protein. The Nanos protein in Drosophila is required for correct morphogenesis in the Drosophila embryo. Translation of the Nanos mRNA is repressed in the bulk cytoplasm and activated in the posterior region. The translation control element (TCE) in the 3'UTR forms a Y-shaped secondary structure, part of which is recognised by the Smaug protein and leads to translational repression.

CUG triplet repeat, RNA binding protein 1, also known as CUGBP1, is a protein which in humans is encoded by the CUGBP1 gene.
Maternal to zygotic transition is the stage in embryonic development during which development comes under the exclusive control of the zygotic genome rather than the maternal (egg) genome. The egg contains stored maternal genetic material mRNA which controls embryo development until the onset of MZT. After MZT the diploid embryo takes over genetic control. This requires both zygotic genome activation (ZGA) and degradation of maternal products. This process is important because it is the first time that the new embryonic genome is utilized and the paternal and maternal genomes are used in combination. The zygotic genome now drives embryo development.

In molecular biology, miR-184 microRNA is a short non-coding RNA molecule. MicroRNAs (miRNAs) function as posttranscriptional regulators of expression levels of other genes by several mechanisms. Several targets for miR-184 have been described, including that of mediators of neurological development, apoptosis and it has been suggested that miR-184 plays an essential role in development.

In molecular biology, the protein domain Sterile alpha motif is a putative protein interaction module present in a wide variety of proteins involved in many biological processes. The SAM domain that spreads over around 70 residues is found in diverse eukaryotic organisms. SAM domains have been shown to homo- and hetero-oligomerise, forming multiple self-association architectures and also binding to various non-SAM domain-containing proteins, nevertheless with a low affinity constant.
In molecular biology mir-430 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
Norbert Perrimon is a geneticist and developmental biologist at Harvard Medical School. He is known for developing a number of techniques for use of Drosophila, as well as specific substantive contributions to signal transduction and developmental biology. Perrimon co-developed the GAL4/UAS system method, described as “a fly geneticist's Swiss army knife”, with Andrea Brand to control gene expression. With Tze-bin Chou he developed the FLP-FRT DFS method to analyze the maternal effect of zygotic lethal mutations. With Jianquan Ni, he developed and improved methods for in vivo RNAi. His lab has pioneered high-throughput whole-genome RNAi screening.

Bicoid is a maternal effect gene whose protein concentration gradient patterns the anterior-posterior (A-P) axis during Drosophila embryogenesis. Bicoid was the first protein demonstrated to act as a morphogen. Although Bicoid is important for the development of Drosophila and other higher dipterans, it is absent from most other insects, where its role is accomplished by other genes.
Ruth Lehmann is a developmental and cell biologist. She is the Director of the Whitehead Institute for Biomedical Research, succeeding David Page. She previously was affiliated with the New York University School of Medicine, where she was the Director of the Skirball Institute of Biomolecular Medicine, the Laura and Isaac Perlmutter Professor of Cell Biology, and the Chair of the Department of Cell Biology. Her research focuses on germ cells and embryogenesis.
Antonio Jesus Giraldez is a Spanish developmental biologist and RNA researcher at Yale University School of Medicine, where he serves as chair of the department of genetics and Fergus F. Wallace Professor of Genetics. He is also affiliated with the Yale Cancer Center and the Yale Stem Cell Center.

Howard David Lipshitz is an American and Canadian biologist who does genetic research on the fruit fly, Drosophila.
Elizabeth Gavis is an American biologist who is the Damon B. Pfeiffer Professor of Life Sciences, at Princeton University. Davis served as the President of the North American Drosophila Board of Directors in 2011.









