
Meiosis is a special type of cell division of germ cells and apicomplexans in sexually-reproducing organisms that produces the gametes, the sperm or egg cells. It involves two rounds of division that ultimately result in four cells, each with only one copy of each chromosome (haploid). Additionally, prior to the division, genetic material from the paternal and maternal copies of each chromosome is crossed over, creating new combinations of code on each chromosome. Later on, during fertilisation, the haploid cells produced by meiosis from a male and a female will fuse to create a zygote, a cell with two copies of each chromosome again.

Chromosomal crossover, or crossing over, is the exchange of genetic material during sexual reproduction between two homologous chromosomes' non-sister chromatids that results in recombinant chromosomes. It is one of the final phases of genetic recombination, which occurs in the pachytene stage of prophase I of meiosis during a process called synapsis. Synapsis begins before the synaptonemal complex develops and is not completed until near the end of prophase I. Crossover usually occurs when matching regions on matching chromosomes break and then reconnect to the other chromosome.

A heteroduplex is a double-stranded (duplex) molecule of nucleic acid originated through the genetic recombination of single complementary strands derived from different sources, such as from different homologous chromosomes or even from different organisms.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids.
The pachytene stage, also known as pachynema, is the third stage of prophase I during meiosis, the specialized cell division that reduces chromosome number by half to produce haploid gametes. It follows the zygotene stage.
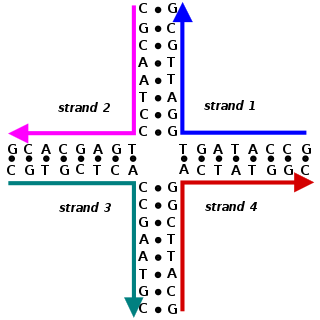
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the junction. The structure is named after Robin Holliday, the molecular biologist who proposed its existence in 1964.

Sister chromatid exchange (SCE) is the exchange of genetic material between two identical sister chromatids.
Chromosome segregation is the process in eukaryotes by which two sister chromatids formed as a consequence of DNA replication, or paired homologous chromosomes, separate from each other and migrate to opposite poles of the nucleus. This segregation process occurs during both mitosis and meiosis. Chromosome segregation also occurs in prokaryotes. However, in contrast to eukaryotic chromosome segregation, replication and segregation are not temporally separated. Instead segregation occurs progressively following replication.

Cell cycle checkpoint protein RAD17 is a protein that in humans is encoded by the RAD17 gene.

Cell cycle checkpoint protein RAD1 is a protein that in humans is encoded by the RAD1 gene.

Exonuclease 1 is an enzyme that in humans is encoded by the EXO1 gene.

MutS protein homolog 5 is a protein that in humans is encoded by the MSH5 gene.
DNA topoisomerase 3-alpha is an enzyme that in humans is encoded by the TOP3A gene.

Meiotic recombination protein DMC1/LIM15 homolog is a protein that in humans is encoded by the DMC1 gene.

MutS protein homolog 4 is a protein that in humans is encoded by the MSH4 gene.

Crossover junction endonuclease MUS81 is an enzyme that in humans is encoded by the MUS81 gene.

DNA mismatch repair protein Mlh3 is a protein that in humans is encoded by the MLH3 gene.
Sgs1, also known as slow growth suppressor 1, is a DNA helicase protein found in Saccharomyces cerevisiae. It is a homolog of the bacterial RecQ helicase. Like the other members of the RecQ helicase family, Sgs1 is important for DNA repair. In particular, Sgs1 collaborates with other proteins to repair double-strand breaks during homologous recombination in eukaryotes.

The meiotic recombination checkpoint monitors meiotic recombination during meiosis, and blocks the entry into metaphase I if recombination is not efficiently processed.

Synthesis-dependent strand annealing (SDSA) is a major mechanism of homology-directed repair of DNA double-strand breaks (DSBs). Although many of the features of SDSA were first suggested in 1976, the double-Holliday junction model proposed in 1983 was favored by many researchers. In 1994, studies of double-strand gap repair in Drosophila were found to be incompatible with the double-Holliday junction model, leading researchers to propose a model they called synthesis-dependent strand annealing. Subsequent studies of meiotic recombination in S. cerevisiae found that non-crossover products appear earlier than double-Holliday junctions or crossover products, challenging the previous notion that both crossover and non-crossover products are produced by double-Holliday junctions and leading the authors to propose that non-crossover products are generated through SDSA.


















