
Bacillus is a genus of Gram-positive, rod-shaped bacteria, a member of the phylum Bacillota, with 266 named species. The term is also used to describe the shape (rod) of other so-shaped bacteria; and the plural Bacilli is the name of the class of bacteria to which this genus belongs. Bacillus species can be either obligate aerobes which are dependent on oxygen, or facultative anaerobes which can survive in the absence of oxygen. Cultured Bacillus species test positive for the enzyme catalase if oxygen has been used or is present.

Bacillus cereus is a Gram-positive rod-shaped bacterium commonly found in soil, food, and marine sponges. The specific name, cereus, meaning "waxy" in Latin, refers to the appearance of colonies grown on blood agar. Some strains are harmful to humans and cause foodborne illness due to their spore-forming nature, while other strains can be beneficial as probiotics for animals, and even exhibit mutualism with certain plants. B. cereus bacteria may be aerobes or facultative anaerobes, and like other members of the genus Bacillus, can produce protective endospores. They have a wide range of virulence factors, including phospholipase C, cereulide, sphingomyelinase, metalloproteases, and cytotoxin K, many of which are regulated via quorum sensing. B. cereus strains exhibit flagellar motility.
Autolysins are endogenous lytic enzymes that break down the peptidoglycan components of biological cells which enables the separation of daughter cells following cell division. They are involved in cell growth, cell wall metabolism, cell division and separation, as well as peptidoglycan turnover and have similar functions to lysozymes.

An endospore is a dormant, tough, and non-reproductive structure produced by some bacteria in the phylum Bacillota. The name "endospore" is suggestive of a spore or seed-like form, but it is not a true spore. It is a stripped-down, dormant form to which the bacterium can reduce itself. Endospore formation is usually triggered by a lack of nutrients, and usually occurs in gram-positive bacteria. In endospore formation, the bacterium divides within its cell wall, and one side then engulfs the other. Endospores enable bacteria to lie dormant for extended periods, even centuries. There are many reports of spores remaining viable over 10,000 years, and revival of spores millions of years old has been claimed. There is one report of viable spores of Bacillus marismortui in salt crystals approximately 25 million years old. When the environment becomes more favorable, the endospore can reactivate itself into a vegetative state. Most types of bacteria cannot change to the endospore form. Examples of bacterial species that can form endospores include Bacillus cereus, Bacillus anthracis, Bacillus thuringiensis, Clostridium botulinum, and Clostridium tetani. Endospore formation is not found among Archaea.

Dormancy is a period in an organism's life cycle when growth, development, and physical activity are temporarily stopped. This minimizes metabolic activity and therefore helps an organism to conserve energy. Dormancy tends to be closely associated with environmental conditions. Organisms can synchronize entry to a dormant phase with their environment through predictive or consequential means. Predictive dormancy occurs when an organism enters a dormant phase before the onset of adverse conditions. For example, photoperiod and decreasing temperature are used by many plants to predict the onset of winter. Consequential dormancy occurs when organisms enter a dormant phase after adverse conditions have arisen. This is commonly found in areas with an unpredictable climate. While very sudden changes in conditions may lead to a high mortality rate among animals relying on consequential dormancy, its use can be advantageous, as organisms remain active longer and are therefore able to make greater use of available resources.

In molecular biology and genetics, transformation is the genetic alteration of a cell resulting from the direct uptake and incorporation of exogenous genetic material from its surroundings through the cell membrane(s). For transformation to take place, the recipient bacterium must be in a state of competence, which might occur in nature as a time-limited response to environmental conditions such as starvation and cell density, and may also be induced in a laboratory.

Bacillus subtilis, known also as the hay bacillus or grass bacillus, is a gram-positive, catalase-positive bacterium, found in soil and the gastrointestinal tract of ruminants, humans and marine sponges. As a member of the genus Bacillus, B. subtilis is rod-shaped, and can form a tough, protective endospore, allowing it to tolerate extreme environmental conditions. B. subtilis has historically been classified as an obligate aerobe, though evidence exists that it is a facultative anaerobe. B. subtilis is considered the best studied Gram-positive bacterium and a model organism to study bacterial chromosome replication and cell differentiation. It is one of the bacterial champions in secreted enzyme production and used on an industrial scale by biotechnology companies.
Candidatus Epulopiscium is a genus of Gram-positive bacteria that have a symbiotic relationship with surgeonfish. These bacteria are known for their unusually large size, many ranging from 0.2 - 0.7 mm in length. Until the discovery of Thiomargarita namibiensis in 1999, Epulonipiscium species were thought to be the largest bacteria. They are still the largest known heterotrophic bacteria.

Lysogeny, or the lysogenic cycle, is one of two cycles of viral reproduction. Lysogeny is characterized by integration of the bacteriophage nucleic acid into the host bacterium's genome or formation of a circular replicon in the bacterial cytoplasm. In this condition the bacterium continues to live and reproduce normally, while the bacteriophage lies in a dormant state in the host cell. The genetic material of the bacteriophage, called a prophage, can be transmitted to daughter cells at each subsequent cell division, and later events can release it, causing proliferation of new phages via the lytic cycle.

In microbiology, genetics, cell biology, and molecular biology, competence is the ability of a cell to alter its genetics by taking up extracellular DNA from its environment through a process called transformation. Competence can be differentiated between natural competence and induced or artificial competence. Natural competence is a genetically specified ability of bacteria that occurs under natural conditions as well as in the laboratory. Artificial competence arises when cells in laboratory cultures are treated to make them transiently permeable to DNA. Competence allows for rapid adaptation and DNA repair of the cell.

Myxococcus xanthus is a gram-negative, bacillus species of myxobacteria that is typically found in the top-most layer of soil. These bacteria lack flagella; rather, they use pili for motility. M. xanthus is well-known for its predatory behavior on other microorganisms. These bacteria source carbon from lipids rather than sugars. They exhibit various forms of self-organizing behavior in response to environmental cues. Under normal conditions with abundant food, they exist as predatory, saprophytic single-species biofilm called a swarm, highlighting the importance of intercellular communication for these bacteria. Under starvation conditions, they undergo a multicellular development cycle.
In biology, an autoinducer is a signaling molecule that enables detection and response to changes in the population density of bacterial cells. Synthesized when a bacterium reproduces, autoinducers pass outside the bacterium and into the surrounding medium. They are a key component of the phenomenon of quorum sensing: as the density of quorum-sensing bacterial cells increases, so does the concentration of the autoinducer. A bacterium’s detection of an autoinducer above some minimum threshold triggers altered gene expression.
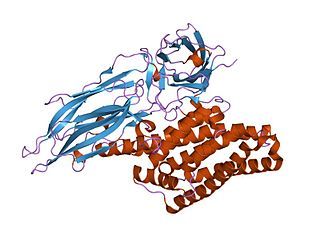
Delta endotoxins (δ-endotoxins) are a family of pore-forming toxins produced by Bacillus thuringiensis species of bacteria. They are useful for their insecticidal action and are the primary toxin produced by the genetically modified (GM) Bt maize/corn and other GM crops. During spore formation the bacteria produce crystals of such proteins that are also known as parasporal bodies, next to the endospores; as a result some members are known as a parasporin. The Cyt (cytolytic) toxin group is another group of delta-endotoxins formed in the cytoplasm. VIP toxins are formed at other stages of the life cycle.
Dictyostelium discoideum is a species of soil-dwelling amoeba belonging to the phylum Amoebozoa, infraphylum Mycetozoa. Commonly referred to as slime mold, D. discoideum is a eukaryote that transitions from a collection of unicellular amoebae into a multicellular slug and then into a fruiting body within its lifetime. Its unique asexual life cycle consists of four stages: vegetative, aggregation, migration, and culmination. The life cycle of D. discoideum is relatively short, which allows for timely viewing of all stages. The cells involved in the life cycle undergo movement, chemical signaling, and development, which are applicable to human cancer research. The simplicity of its life cycle makes D. discoideum a valuable model organism to study genetic, cellular, and biochemical processes in other organisms.
Spore photoproduct lyase is a radical SAM enzyme that repairs DNA cross linking of thymine bases caused by UV-radiation. There are several types of thymine cross linking, but SPL specifically targets 5-thyminyl-5,6-dihydrothymine, which is also called spore photoproduct (SP). Spore photoproduct is the predominant type of thymine crosslinking in germinating endospores, which is why SPL is unique to organisms that produce endospores, such as Bacillus subtilis. Other types of thymine crosslinking, such as cyclobutane pyrimidine dimers (CPD) and pyrimidine (6-4) pyrimidone photoproducts (6-4PPs), are less commonly formed in endospores. These differences in DNA crosslinking are a function of differing DNA structure. Spore genomic DNA features many DNA binding proteins called small acid soluble proteins, which changes the DNA from the traditional B-form conformation to an A-form conformation. This difference in conformation is believed to be the reason why dormant spores predominantly accumulate SP in response to UV-radiation, rather than other forms of cross linking. Spores cannot repair cross-linking while dormant, instead the SPs are repaired during germination to allow the vegetative cell to function normally. When not repaired, spore photoproduct and other types of crosslinking can cause mutations by blocking transcription and replication past the point of the crosslinking. The repair mechanism utilizing spore photoproduct lyase is one of the reasons for the resilience of certain bacterial spores.

Bacillus anthracis is a gram-positive and rod-shaped bacterium that causes anthrax, a deadly disease to livestock and, occasionally, to humans. It is the only permanent (obligate) pathogen within the genus Bacillus. Its infection is a type of zoonosis, as it is transmitted from animals to humans. It was discovered by a German physician Robert Koch in 1876, and became the first bacterium to be experimentally shown as a pathogen. The discovery was also the first scientific evidence for the germ theory of diseases.

In molecular biology, the protein domain Sda is short for suppressor of dnaA or otherwise known as sporulation inhibitor A. It is found only in bacteria. This protein domain is highly important to cell survival. When starved of nutrients, the cell is under extreme stress so undergoes a series of reactions to increase the chances of survival. One method is to form endospores which can withstand a large amount of environmental pressure. Sda protein domain is a checkpoint which prevents the formation of spores. The Sda domain affects cell signalling. It prevents the cell communicating the stress that it is under, which is crucial if the cell is to survive.

Endospore staining is a technique used in bacteriology to identify the presence of endospores in a bacterial sample. Within bacteria, endospores are protective structures used to survive extreme conditions, including high temperatures making them highly resistant to chemicals. Endospores contain little or no ATP which indicates how dormant they can be. Endospores contain a tough outer coating made up of keratin which protects them from nucleic DNA as well as other adaptations. Endospores are able to regerminate into vegetative cells, which provides a protective nature that makes them difficult to stain using normal techniques such as simple staining and gram staining. Special techniques for endospore staining include the Schaeffer–Fulton stain and the Moeller stain.
The Monovalent Cation:Proton Antiporter-2 (CPA2) Family is a moderately large family of transporters belonging to the CPA superfamily. Members of the CPA2 family have been found in bacteria, archaea and eukaryotes. The proteins of the CPA2 family consist of between 333 and 900 amino acyl residues and exhibit 10-14 transmembrane α-helical spanners (TMSs).

The Phosphate (Pho) regulon is a regulatory mechanism used for the conservation and management of inorganic phosphate within the cell. It was first discovered in Escherichia coli as an operating system for the bacterial strain, and was later identified in other species. The Pho system is composed of various components including extracellular enzymes and transporters that are capable of phosphate assimilation in addition to extracting inorganic phosphate from organic sources. This is an essential process since phosphate plays an important role in cellular membranes, genetic expression, and metabolism within the cell. Under low nutrient availability, the Pho regulon helps the cell survive and thrive despite a depletion of phosphate within the environment. When this occurs, phosphate starvation-inducible (psi) genes activate other proteins that aid in the transport of inorganic phosphate.
















