Related Research Articles

Thiamine, also known as thiamin and vitamin B1, is a vitamin, an essential micronutrient for humans and animals. It is found in food and commercially synthesized to be a dietary supplement or medication. Phosphorylated forms of thiamine are required for some metabolic reactions, including the breakdown of glucose and amino acids.

In molecular biology, a riboswitch is a regulatory segment of a messenger RNA molecule that binds a small molecule, resulting in a change in production of the proteins encoded by the mRNA. Thus, an mRNA that contains a riboswitch is directly involved in regulating its own activity, in response to the concentrations of its effector molecule. The discovery that modern organisms use RNA to bind small molecules, and discriminate against closely related analogs, expanded the known natural capabilities of RNA beyond its ability to code for proteins, catalyze reactions, or to bind other RNA or protein macromolecules.
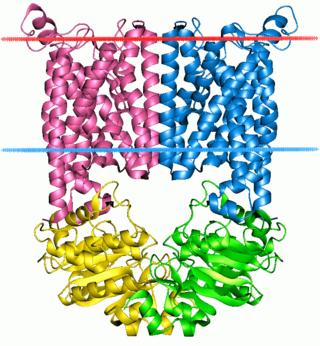
The ATP-binding cassette transporters are a transport system superfamily that is one of the largest and possibly one of the oldest gene families. It is represented in all extant phyla, from prokaryotes to humans. ABC transporters belong to translocases.
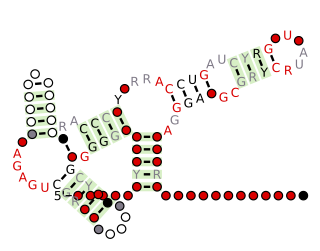
The TPP riboswitch, also known as the THI element and Thi-box riboswitch, is a highly conserved RNA secondary structure. It serves as a riboswitch that binds thiamine pyrophosphate (TPP) directly and modulates gene expression through a variety of mechanisms in archaea, bacteria and eukaryotes. TPP is the active form of thiamine (vitamin B1), an essential coenzyme synthesised by coupling of pyrimidine and thiazole moieties in bacteria. The THI element is an extension of a previously detected thiamin-regulatory element, the thi box, there is considerable variability in the predicted length and structures of the additional and facultative stem-loops represented in dark blue in the secondary structure diagram Analysis of operon structures has identified a large number of new candidate thiamin-regulated genes, mostly transporters, in various prokaryotic organisms. The x-ray crystal structure of the TPP riboswitch aptamer has been solved.

Thiamine transporter 1, also known as thiamine carrier 1 (TC1) or solute carrier family 19 member 2 (SLC19A2) is a protein that in humans is encoded by the SLC19A2 gene. SLC19A2 is a thiamine transporter. Mutations in this gene cause thiamine-responsive megaloblastic anemia syndrome (TRMA), which is an autosomal recessive disorder characterized by diabetes mellitus, megaloblastic anemia and sensorineural deafness.

Thiamine transporter 2 (ThTr-2), also known as solute carrier family 19 member 3, is a protein that in humans is encoded by the SLC19A3 gene. SLC19A3 is a thiamine transporter.

ATP-binding cassette transporter ABCA1, also known as the cholesterol efflux regulatory protein (CERP) is a protein which in humans is encoded by the ABCA1 gene. This transporter is a major regulator of cellular cholesterol and phospholipid homeostasis.
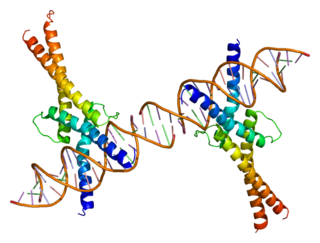
Sterol regulatory element-binding transcription factor 1 (SREBF1) also known as sterol regulatory element-binding protein 1 (SREBP-1) is a protein that in humans is encoded by the SREBF1 gene.

Sterol regulatory element-binding protein 2 (SREBP-2) also known as sterol regulatory element binding transcription factor 2 (SREBF2) is a protein that in humans is encoded by the SREBF2 gene.

Activating transcription factor 4 , also known as ATF4, is a protein that in humans is encoded by the ATF4 gene.
Autoinducers are signaling molecules that are produced in response to changes in cell-population density. As the density of quorum sensing bacterial cells increases so does the concentration of the autoinducer. Detection of signal molecules by bacteria acts as stimulation which leads to altered gene expression once the minimal threshold is reached. Quorum sensing is a phenomenon that allows both Gram-negative and Gram-positive bacteria to sense one another and to regulate a wide variety of physiological activities. Such activities include symbiosis, virulence, motility, antibiotic production, and biofilm formation. Autoinducers come in a number of different forms depending on the species, but the effect that they have is similar in many cases. Autoinducers allow bacteria to communicate both within and between different species. This communication alters gene expression and allows bacteria to mount coordinated responses to their environments, in a manner that is comparable to behavior and signaling in higher organisms. Not surprisingly, it has been suggested that quorum sensing may have been an important evolutionary milestone that ultimately gave rise to multicellular life forms.

Nuclear factor of activated T-cells 5, also known as NFAT5 and sometimes TonEBP, is a human gene that encodes a transcription factor that regulates the expression of genes involved in the osmotic stress.

ATP-binding cassette sub-family A member 2 is a protein that in humans is encoded by the ABCA2 gene.

ATP-binding cassette sub-family A member 7 is a protein that in humans is encoded by the ABCA7 gene.
The Magnesium responsive RNA element, not to be confused with the completely distinct M-box riboswitch, is a cis-regulatory element that regulates the expression of the magnesium transporter protein MgtA. It is located in the 5' UTR of this gene. The mechanism for the potential magnesium-sensing capacity of this RNA is still unclear, though a recent report suggests that the RNA element targets the mgtA transcript for degradation by RNase E when cells are grown in high Mg2+ environments.

The fluoride riboswitch is a conserved RNA structure identified by bioinformatics in a wide variety of bacteria and archaea. These RNAs were later shown to function as riboswitches that sense fluoride ions. These "fluoride riboswitches" increase expression of downstream genes when fluoride levels are elevated, and the genes are proposed to help mitigate the toxic effects of very high levels of fluoride.

The glutamine riboswitch is a conserved RNA structure that was predicted by bioinformatics. It is present in a variety of lineages of cyanobacteria, as well as some phages that infect cyanobacteria. It is also found in DNA extracted from uncultivated bacteria living in the ocean that are presumably species of cyanobacteria.
Bacterial small RNAs (bsRNA) are small RNAs produced by bacteria; they are 50- to 500-nucleotide non-coding RNA molecules, highly structured and containing several stem-loops. Numerous sRNAs have been identified using both computational analysis and laboratory-based techniques such as Northern blotting, microarrays and RNA-Seq in a number of bacterial species including Escherichia coli, the model pathogen Salmonella, the nitrogen-fixing alphaproteobacterium Sinorhizobium meliloti, marine cyanobacteria, Francisella tularensis, Streptococcus pyogenes, the pathogen Staphylococcus aureus, and the plant pathogen Xanthomonas oryzae pathovar oryzae. Bacterial sRNAs affect how genes are expressed within bacterial cells via interaction with mRNA or protein, and thus can affect a variety of bacterial functions like metabolism, virulence, environmental stress response, and structure.

miR-33 is a family of microRNA precursors, which are processed by the Dicer enzyme to give mature microRNAs. miR-33 is found in several animal species, including humans. In some species there is a single member of this family which gives the mature product mir-33. In humans there are two members of this family called mir-33a and mir-33b, which are located in intronic regions within two protein-coding genes for Sterol regulatory element-binding proteins respectively.
In bacterial genetics, the mal regulon is a regulon - or group of genes under common regulation - associated with the catabolism of maltose and maltodextrins. The system is especially well characterized in the model organism Escherichia coli, where it is classically described as a group of ten genes in multiple operons whose expression is regulated by a single regulatory protein, malT. MalT binds to maltose or maltodextrin and undergoes a conformational change that allows it to bind DNA at sequences near the promoters of genes required for uptake and catabolism of these sugars. The maltose regulation system in E. coli is a classic example of positive regulation. malT is regulated by catabolite repression via the catabolite activator protein. Genes under the control of malT include ATP-binding cassette transporter components, maltoporin, maltose binding protein, and several enzymes. Other Gram-negative bacteria such as Klebsiella pneumoniae have additional genes under the control of malT.
References
- ↑ Erkens GB, Slotboom DJ (April 2010). "Biochemical characterization of ThiT from Lactococcus lactis: a thiamin transporter with picomolar substrate binding affinity". Biochemistry. 49 (14): 3203–12. doi:10.1021/bi100154r. PMID 20218726.
- ↑ Eitinger T, Rodionov DA, Grote M, Schneider E (January 2011). "Canonical and ECF-type ATP-binding cassette importers in prokaryotes: diversity in modular organization and cellular functions". FEMS Microbiology Reviews. 35 (1): 3–67. doi: 10.1111/j.1574-6976.2010.00230.x . PMID 20497229.