Related Research Articles

The enzyme-linked immunosorbent assay (ELISA) is a commonly used analytical biochemistry assay, first described by Engvall and Perlmann in 1971. The assay uses a solid-phase type of enzyme immunoassay (EIA) to detect the presence of a ligand in a liquid sample using antibodies directed against the protein to be measured. ELISA has been used as a diagnostic tool in medicine, plant pathology, and biotechnology, as well as a quality control check in various industries.
Viral load, also known as viral burden, is a numerical expression of the quantity of virus in a given volume of fluid, including biological and environmental specimens. It is not to be confused with viral titre or viral titer, which depends on the assay. When an assay for measuring the infective virus particle is done, viral titre often refers to the concentration of infectious viral particles, which is different from the total viral particles. Sputum and blood plasma are two bodily fluids from which viral load is measured. As an example of environmental specimens, the viral load of norovirus can be determined from run-off water on garden produce. Norovirus has not only prolonged viral shedding and has the ability to survive in the environment but a minuscule infectious dose is required to produce infection in humans: less than 100 viral particles.

Sputum is mucus that is coughed up from the lower airways. In medicine, sputum samples are usually used for naked eye exam, microbiological investigations of respiratory infections and cytological investigations of respiratory systems. It is critical that the patient not give a specimen that includes any mucoid material from the interior of the nose.

A microbiological culture, or microbial culture, is a method of multiplying microbial organisms by letting them reproduce in predetermined culture medium under controlled laboratory conditions. Microbial cultures are foundational and basic diagnostic methods used as a research tool in molecular biology.

Bacteriological water analysis is a method of analysing water to estimate the numbers of bacteria present and, if needed, to find out what sort of bacteria they are. It represents one aspect of water quality. It is a microbiological analytical procedure which uses samples of water and from these samples determines the concentration of bacteria. It is then possible to draw inferences about the suitability of the water for use from these concentrations. This process is used, for example, to routinely confirm that water is safe for human consumption or that bathing and recreational waters are safe to use.
Plate readers, also known as microplate readers or microplate photometers, are instruments which are used to detect biological, chemical or physical events of samples in microtiter plates. They are widely used in research, drug discovery, bioassay validation, quality control and manufacturing processes in the pharmaceutical and biotechnological industry and academic organizations. Sample reactions can be assayed in 1-1536 well format microtiter plates. The most common microplate format used in academic research laboratories or clinical diagnostic laboratories is 96-well with a typical reaction volume between 100 and 200 µL per well. Higher density microplates are typically used for screening applications, when throughput and assay cost per sample become critical parameters, with a typical assay volume between 5 and 50 µL per well. Common detection modes for microplate assays are absorbance, fluorescence intensity, luminescence, time-resolved fluorescence, and fluorescence polarization.
Serology is the scientific study of serum and other body fluids. In practice, the term usually refers to the diagnostic identification of antibodies in the serum. Such antibodies are typically formed in response to an infection, against other foreign proteins, or to one's own proteins. In either case, the procedure is simple.

The oxidase test is used to determine if an organism possesses the cytochrome oxidase enzyme. The test is used as an aid for the differentiation of Neisseria, Moraxella, Campylobacter and Pasteurella species. It is also used to differentiate pseudomonads from related species..
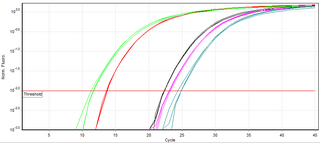
A real-time polymerase chain reaction is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR, not at its end, as in conventional PCR. Real-time PCR can be used quantitatively and semi-quantitatively.
A colony-forming unit is a unit used in microbiology to estimate the number of viable bacteria or fungal cells in a sample. Viable is defined as the ability to multiply via binary fission under the controlled conditions. Counting with colony-forming units requires culturing the microbes and counts only viable cells, in contrast with microscopic examination which counts all cells, living or dead. The visual appearance of a colony in a cell culture requires significant growth, and when counting colonies it is uncertain if the colony arose from one cell or a group of cells. Expressing results as colony-forming units reflects this uncertainty.
Indicator bacteria are types of bacteria used to detect and estimate the level of fecal contamination of water. They are not dangerous to human health but are used to indicate the presence of a health risk.
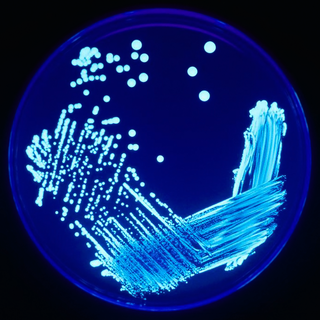
In microbiology, streaking is a technique used to isolate a pure strain from a single species of microorganism, often bacteria. Samples can then be taken from the resulting colonies and a microbiological culture can be grown on a new plate so that the organism can be identified, studied, or tested.
Indoor bioaerosol is bioaerosol in an indoor environment. Bioaerosols are natural or artificial particles of biological origin suspended in the air. These particles are also referred to as organic dust. Bioaerosols may consist of bacteria, fungi, viruses, microbial toxins, pollen, plant fibers, etc. Size of bioaerosol particles varies from below 1 µm to 100 µm in aerodynamic diameter; viable bioaerosol particles can be suspended in air as single cells or aggregates of microorganism as small as 1–10 µm in size. Since bioaerosols are potentially related to various human health effects and the indoor environment provides a unique exposure situation, concerns about indoor bioaerosols have increased over the last decade.
Mycobacteria Growth Indicator Tube (MGIT) is intended for the detection and recovery of mycobacteria. The MGIT Mycobacteria Growth Indicator Tube contains 7 mL of modified Middlebrook 7H9 Broth base. The complete medium, with OADC enrichment and PANTA antibiotic mixture, is one of the most commonly used liquid media for the cultivation of mycobacteria.
Virus quantification involves counting the number of viruses in a specific volume to determine the virus concentration. It is utilized in both research and development (R&D) in commercial and academic laboratories as well as production situations where the quantity of virus at various steps is an important variable. For example, the production of viral vaccines, recombinant proteins using viral vectors and viral antigens all require virus quantification to continually adapt and monitor the process in order to optimize production yields and respond to ever changing demands and applications. Examples of specific instances where known viruses need to be quantified include clone screening, multiplicity of infection (MOI) optimization and adaptation of methods to cell culture. This page discusses various techniques currently used to quantify viruses in liquid samples. These methods are separated into two categories, traditional vs. modern methods. Traditional methods are industry-standard methods that have been used for decades but are generally slow and labor-intensive. Modern methods are relatively new commercially available products and kits that greatly reduce quantification time. This is not meant to be an exhaustive review of all potential methods, but rather a representative cross-section of traditional methods and new, commercially available methods. While other published methods may exist for virus quantification, non-commercial methods are not discussed here.
Cell counting is any of various methods for the counting or similar quantification of cells in the life sciences, including medical diagnosis and treatment. It is an important subset of cytometry, with applications in research and clinical practice. For example, the complete blood count can help a physician to determine why a patient feels unwell and what to do to help. Cell counts within liquid media are usually expressed as a number of cells per unit of volume, thus expressing a concentration.
Bioburden is normally defined as the number of bacteria living on a surface that has not been sterilized.
Spot analysis, spot test analysis, or spot test is a chemical test, a simple and efficient technique where analytic assays are executed in only one, or a few drops, of a chemical solution, preferably in a great piece of filter paper, without using any sophisticated instrumentation. The development and popularization of the test is credited to Fritz Feigl.

A spiral plater is an instrument used to dispense a liquid sample onto a Petri dish in a spiral pattern. Commonly used as part of a CFU count procedure for the purpose of determining the number of microbes in the sample. In this setting, after spiral plating, the Petri dish is incubated for several hours after which the number of colony forming microbes (CFU) is determined. Spiral platers are also used for research, clinical diagnostics and as a method for covering a Petri dish with bacteria before placing antibiotic discs for AST.
Impedance microbiology is a microbiological technique used to measure the microbial number density of a sample by monitoring the electrical parameters of the growth medium. The ability of microbial metabolism to change the electrical conductivity of the growth medium was discovered by Stewart and further studied by other scientists such as Oker-Blom, Parson and Allison in the first half of 20th century. However, it was only in the late 1970s that, thanks to computer-controlled systems used to monitor impedance, the technique showed its full potential, as discussed in the works of Fistenberg-Eden & Eden, Ur & Brown and Cady.
References
- ↑ Massicotte, Richard; Mafu, Akier A; Ahmad, Darakhshan; Deshaies, Francis; Pichette, Gilbert; Belhumeur, Pierre (2017). "Comparison between Flow Cytometry and Traditional Culture Methods for Efficacy Assessment of Six Disinfectant Agents against Nosocomial Bacterial Species". Frontiers in Microbiology. 8: 112. doi: 10.3389/fmicb.2017.00112 . PMC 5289957 . PMID 28217115.