
Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that help such reactions are called proteases.

Ubiquitin is a small regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ubiquitously. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A.

A ubiquitin ligase is a protein that recruits an E2 ubiquitin-conjugating enzyme that has been loaded with ubiquitin, recognizes a protein substrate, and assists or directly catalyzes the transfer of ubiquitin from the E2 to the protein substrate. In simple and more general terms, the ligase enables movement of ubiquitin from a ubiquitin carrier to another thing by some mechanism. The ubiquitin, once it reaches its destination, ends up being attached by an isopeptide bond to a lysine residue, which is part of the target protein. E3 ligases interact with both the target protein and the E2 enzyme, and so impart substrate specificity to the E2. Commonly, E3s polyubiquitinate their substrate with Lys48-linked chains of ubiquitin, targeting the substrate for destruction by the proteasome. However, many other types of linkages are possible and alter a protein's activity, interactions, or localization. Ubiquitination by E3 ligases regulates diverse areas such as cell trafficking, DNA repair, and signaling and is of profound importance in cell biology. E3 ligases are also key players in cell cycle control, mediating the degradation of cyclins, as well as cyclin dependent kinase inhibitor proteins. The human genome encodes over 600 putative E3 ligases, allowing for tremendous diversity in substrates.

Ubiquitin-like modifier activating enzyme 1 (UBA1) is an enzyme which in humans is encoded by the UBA1 gene. UBA1 participates in ubiquitination and the NEDD8 pathway for protein folding and degradation, among many other biological processes. This protein has been linked to X-linked spinal muscular atrophy type 2, neurodegenerative diseases, and cancers.

Deubiquitinating enzymes (DUBs), also known as deubiquitinating peptidases, deubiquitinating isopeptidases, deubiquitinases, ubiquitin proteases, ubiquitin hydrolases, or ubiquitin isopeptidases, are a large group of proteases that cleave ubiquitin from proteins. Ubiquitin is attached to proteins in order to regulate the degradation of proteins via the proteasome and lysosome; coordinate the cellular localisation of proteins; activate and inactivate proteins; and modulate protein-protein interactions. DUBs can reverse these effects by cleaving the peptide or isopeptide bond between ubiquitin and its substrate protein. In humans there are nearly 100 DUB genes, which can be classified into two main classes: cysteine proteases and metalloproteases. The cysteine proteases comprise ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), Machado-Josephin domain proteases (MJDs) and ovarian tumour proteases (OTU). The metalloprotease group contains only the Jab1/Mov34/Mpr1 Pad1 N-terminal+ (MPN+) (JAMM) domain proteases.
In molecular biology, SUMOproteins are a family of small proteins that are covalently attached to and detached from other proteins in cells to modify their function. This process is called SUMOylation. SUMOylation is a post-translational modification involved in various cellular processes, such as nuclear-cytosolic transport, transcriptional regulation, apoptosis, protein stability, response to stress, and progression through the cell cycle.

An isopeptide bond is a type of amide bond formed between a carboxyl group of one amino acid and an amino group of another. An isopeptide bond is the linkage between the side chain amino or carboxyl group of one amino acid to the α-carboxyl, α-amino group, or the side chain of another amino acid. In a typical peptide bond, also known as eupeptide bond, the amide bond always forms between the α-carboxyl group of one amino acid and the α-amino group of the second amino acid. Isopeptide bonds are rarer than regular peptide bonds. Isopeptide bonds lead to branching in the primary sequence of a protein. Proteins formed from normal peptide bonds typically have a linear primary sequence.

Ubiquitin-activating enzymes, also known as E1 enzymes, catalyze the first step in the ubiquitination reaction, which can target a protein for degradation via a proteasome. This covalent bond of ubiquitin or ubiquitin-like proteins to targeted proteins is a major mechanism for regulating protein function in eukaryotic organisms. Many processes such as cell division, immune responses and embryonic development are also regulated by post-translational modification by ubiquitin and ubiquitin-like proteins.

Valosin-containing protein (VCP) or transitional endoplasmic reticulum ATPase also known as p97 in mammals and CDC48 in S. cerevisiae, is an enzyme that in humans is encoded by the VCP gene. The TER ATPase is an ATPase enzyme present in all eukaryotes and archaebacteria. Its main function is to segregate protein molecules from large cellular structures such as protein assemblies, organelle membranes and chromatin, and thus facilitate the degradation of released polypeptides by the multi-subunit protease proteasome.

Ubiquitin-conjugating enzyme E2 L3 (UBE2L3), also called UBCH7, is a protein that in humans is encoded by the UBE2L3 gene. As an E2 enzyme, UBE2L3 participates in ubiquitination to target proteins for degradation. The role of UBE2L3 in the ubiquitination of the NF-κB precursor implicated it in various major autoimmune diseases, including rheumatoid arthritis (RA), celiac disease, Crohn's disease (CD), and systemic lupus erythematosus.

Ubiquitin-conjugating enzyme E2 variant 2 is a protein that in humans is encoded by the UBE2V2 gene. Ubiquitin-conjugating enzyme E2 variant proteins constitute a distinct subfamily within the E2 protein family.

Ubiquitin carboxyl-terminal hydrolase or Ubiquitin specific protease 11 is an enzyme that in humans is encoded by the USP11 gene. USP11 belongs to the Ubiquitin specific proteases family (USPs) which is a sub-family of the Deubiquitinating enzymes (DUBs).USPs are multiple domain proteases and belong to the C19 cysteine proteases sub‒family. Depending on their domain architecture and position there is different homology between the various members. Generally the largest domain is the catalytic domain which harbours the three residue catalytic triad that is included inside conserved motifs. The catalytic domain also contains sequences that are not related with the catalysis function and their role is mostly not clearly understood at present, the length of these sequences varies for each USP and therefore the length of the whole catalytic domain can range from approximately 295 to 850 amino acids. Particular sequences inside the catalytic domain or at the N‒terminus of some USPs have been characterised as UBL and DUSP domains respectively. In some cases, regarding the UBL domains, it has been reported to have a catalysis enhancing function as in the case of USP7. In addition, a so‒called DU domain module is the combination of a DUSP domain followed by a UBL domain separated by a linker and is found in USP11 as well as in USP15 and USP4.

The BTB/POZ domain is a structural domain found in proteins across the domain Eukarya. Given its prevalence in eukaryotes and its absence in Archaea and bacteria, it likely arose after the origin of eukaryotes. While primarily a protein-protein interaction domain, some BTB domains have additional functionality in transcriptional regulation, cytoskeletal mobility, protein ubiquitination and degradation, and ion channel formation and operation. BTB domains have traditionally been classified by the other structural features present in the protein.
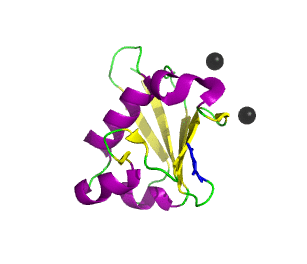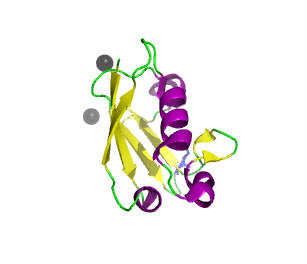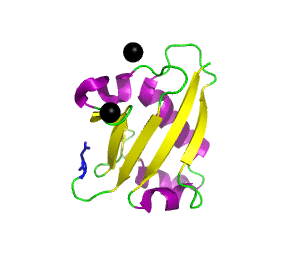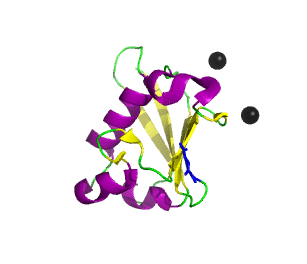
Ubiquitin-related modifier-1 (URM1) is a ubiquitin-like protein that modifies proteins in the yeast ubiquitin-like urmylation pathway. Structural comparisons and phylogenetic analysis of the ubiquitin superfamily has indicated that Urm1 has the most conserved structural and sequence features of the common ancestor of the entire superfamily.

UBX domain protein 6 is a protein in humans that is encoded by the UBXN6 gene.

Ubiquitin-associated (UBA) domains are protein domains that non-covalently interact with ubiquitin through protein-protein interactions. Ubiquitin is a small protein that is covalently linked to other proteins as part of intracellular signaling pathways, often as a signal for protein degradation. UBA domains are among the most common ubiquitin-binding domains.
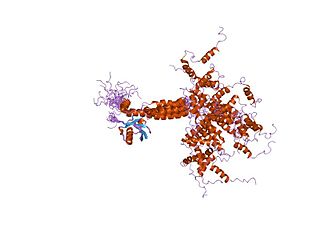
In molecular biology, the Ubiquitin-Interacting Motif (UIM), or 'LALAL-motif', is a sequence motif of about 20 amino acid residues, which was first described in the 26S proteasome subunit PSD4/RPN-10 that is known to recognise ubiquitin. In addition, the UIM is found, often in tandem or triplet arrays, in a variety of proteins either involved in ubiquitination and ubiquitin metabolism, or known to interact with ubiquitin-like modifiers. Among the UIM proteins are two different subgroups of the UBP family of deubiquitinating enzymes, one F-box protein, one family of HECT-containing ubiquitin-ligases (E3s) from plants, and several proteins containing ubiquitin-associated UBA and/or UBX domains. In most of these proteins, the UIM occurs in multiple copies and in association with other domains such as UBA, UBX, ENTH domain, EH, VHS, SH3 domain, HECT, VWFA, EF-hand calcium-binding, WD-40, F-box, LIM, protein kinase, ankyrin, PX, phosphatidylinositol 3- and 4-kinase, C2 domain, OTU, DnaJ domain, RING-finger or FYVE-finger. UIMs have been shown to bind ubiquitin and to serve as a specific targeting signal important for monoubiquitination. Thus, UIMs may have several functions in ubiquitin metabolism each of which may require different numbers of UIMs.

In molecular biology, TUG-UBL1 refers to a protein that regulates a glucose transporter called GLUT4. TUG-UBL1 is an acronym for Tether containing UBX domain for GLUT4-Ubiquitin Like 1, this is encoded for by the gene, ASPSCR1.

Ubiquitin-like proteins (UBLs) are a family of small proteins involved in post-translational modification of other proteins in a cell, usually with a regulatory function. The UBL protein family derives its name from the first member of the class to be discovered, ubiquitin (Ub), best known for its role in regulating protein degradation through covalent modification of other proteins. Following the discovery of ubiquitin, many additional evolutionarily related members of the group were described, involving parallel regulatory processes and similar chemistry. UBLs are involved in a widely varying array of cellular functions including autophagy, protein trafficking, inflammation and immune responses, transcription, DNA repair, RNA splicing, and cellular differentiation.
UBXD8 is a protein in the Ubiquitin regulatory X (UBX) domain-containing protein family. The UBX domain contains many eukaryotic proteins that have similarities in amino acid sequence to the tiny protein modifier ubiquitin. UBXD8 engages in a molecular interaction with p97, a protein that is essential for the degradation of membrane proteins associated with the endoplasmic reticulum (ER) through the proteasome. Ubxd8 possesses a UBA domain, alongside the UBX domain, that could interact with polyubiquitin chains. Additionally, it possesses a UAS domain of undetermined function, and this protein is used as a protein sensor that detects long chain unsaturated fatty acids (FAs), having a vital function in regulating the balance of Fatty Acids within cells to maintain cellular homeostasis.


















