Related Research Articles
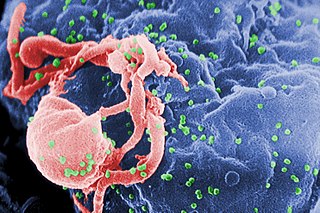
The human immunodeficiency viruses (HIV) are two species of Lentivirus that infect humans. Over time, they cause acquired immunodeficiency syndrome (AIDS), a condition in which progressive failure of the immune system allows life-threatening opportunistic infections and cancers to thrive. Without treatment, average survival time after infection with HIV is estimated to be 9 to 11 years, depending on the HIV subtype.

A retrovirus is a type of virus that inserts a DNA copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. Once inside the host cell's cytoplasm, the virus uses its own reverse transcriptase enzyme to produce DNA from its RNA genome, the reverse of the usual pattern, thus retro (backwards). The new DNA is then incorporated into the host cell genome by an integrase enzyme, at which point the retroviral DNA is referred to as a provirus. The host cell then treats the viral DNA as part of its own genome, transcribing and translating the viral genes along with the cell's own genes, producing the proteins required to assemble new copies of the virus. Many retroviruses cause serious diseases in humans, other mammals, and birds.
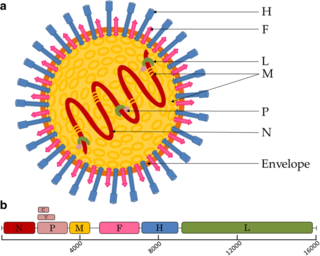
Paramyxoviridae is a family of negative-strand RNA viruses in the order Mononegavirales. Vertebrates serve as natural hosts. Diseases associated with this family include measles, mumps, and respiratory tract infections. The family has four subfamilies, 17 genera, and 78 species, three genera of which are unassigned to a subfamily.

The Epstein–Barr virus (EBV), formally called Human gammaherpesvirus 4, is one of the nine known human herpesvirus types in the herpes family, and is one of the most common viruses in humans. EBV is a double-stranded DNA virus.

A poliovirus, the causative agent of polio, is a serotype of the species Enterovirus C, in the family of Picornaviridae. There are three poliovirus serotypes: types 1, 2, and 3.
Viral evolution is a subfield of evolutionary biology and virology that is specifically concerned with the evolution of viruses. Viruses have short generation times, and many—in particular RNA viruses—have relatively high mutation rates. Although most viral mutations confer no benefit and often even prove deleterious to viruses, the rapid rate of viral mutation combined with natural selection allows viruses to quickly adapt to changes in their host environment. In addition, because viruses typically produce many copies in an infected host, mutated genes can be passed on to many offspring quickly. Although the chance of mutations and evolution can change depending on the type of virus, viruses overall have high chances for mutations.

Hepadnaviridae is a family of viruses. Humans, apes, and birds serve as natural hosts. There are currently 18 species in this family, divided among 5 genera. Its best-known member is hepatitis B virus. Diseases associated with this family include: liver infections, such as hepatitis, hepatocellular carcinomas, and cirrhosis. It is the sole accepted family in the order Blubervirales.

Gammaretrovirus is a genus in the Retroviridae family. Example species are the murine leukemia virus and the feline leukemia virus. They cause various sarcomas, leukemias and immune deficiencies in mammals, reptiles and birds.

Herpesviridae is a large family of DNA viruses that cause infections and certain diseases in animals, including humans. The members of this family are also known as herpesviruses. The family name is derived from the Greek word ἕρπειν, referring to spreading cutaneous lesions, usually involving blisters, seen in flares of herpes simplex 1, herpes simplex 2 and herpes zoster (shingles). In 1971, the International Committee on the Taxonomy of Viruses (ICTV) established Herpesvirus as a genus with 23 viruses among four groups. As of 2020, 115 species are recognized, all but one of which are in one of the three subfamilies. Herpesviruses can cause both latent and lytic infections.

Herpes simplex virus1 and 2, also known by their taxonomical names Human alphaherpesvirus 1 and Human alphaherpesvirus 2, are two members of the human Herpesviridae family, a set of viruses that produce viral infections in the majority of humans. Both HSV-1 and HSV-2 are very common and contagious. They can be spread when an infected person begins shedding the virus.

Duck plague is a worldwide disease caused by Anatid alphaherpesvirus 1 (AnHV-1) of the family Herpesviridae that causes acute disease with high mortality rates in flocks of ducks, geese, and swans. It is spread both vertically and horizontally—through contaminated water and direct contact. Migratory waterfowl are a major factor in the spread of this disease as they are often asymptomatic carriers of disease. The incubation period is three to seven days. Birds as young as one week old can be infected. DEV is not zoonotic.
Simian foamy virus (SFV) is a species of the genus Spumavirus that belongs to the family of Retroviridae. It has been identified in a wide variety of primates, including prosimians, New World and Old World monkeys, as well as apes, and each species has been shown to harbor a unique (species-specific) strain of SFV, including African green monkeys, baboons, macaques, and chimpanzees. As it is related to the more well-known retrovirus human immunodeficiency virus (HIV), its discovery in primates has led to some speculation that HIV may have been spread to the human species in Africa through contact with blood from apes, monkeys, and other primates, most likely through bushmeat-hunting practices.

Gammaherpesvirinae is a subfamily of viruses in the order Herpesvirales and in the family Herpesviridae. Viruses in Gammaherpesvirinae are distinguished by reproducing at a more variable rate than other subfamilies of Herpesviridae. Mammals serve as natural hosts. There are 43 species in this subfamily, divided among 7 genera with three species unassigned to a genus. Diseases associated with this subfamily include: HHV-4: infectious mononucleosis. HHV-8: Kaposi's sarcoma.
Phycodnaviridae is a family of large (100–560 kb) double-stranded DNA viruses that infect marine or freshwater eukaryotic algae. Viruses within this family have a similar morphology, with an icosahedral capsid. As of 2014, there were 33 species in this family, divided among 6 genera. This family belongs to a super-group of large viruses known as nucleocytoplasmic large DNA viruses. Evidence was published in 2014 suggesting that specific strains of Phycodnaviridae might infect humans rather than just algal species, as was previously believed. Most genera under this family enter the host cell by cell receptor endocytosis and replicate in the nucleus. Phycodnaviridae play important ecological roles by regulating the growth and productivity of their algal hosts. Algal species such Heterosigma akashiwo and the genus Chrysochromulina can form dense blooms which can be damaging to fisheries, resulting in losses in the aquaculture industry. Heterosigma akashiwo virus (HaV) has been suggested for use as a microbial agent to prevent the recurrence of toxic red tides produced by this algal species. Phycodnaviridae cause death and lysis of freshwater and marine algal species, liberating organic carbon, nitrogen and phosphorus into the water, providing nutrients for the microbial loop.

Alfalfa mosaic virus (AMV), also known as Lucerne mosaic virus or Potato calico virus, is a worldwide distributed phytopathogen that can lead to necrosis and yellow mosaics on a large variety of plant species, including commercially important crops. It is the only Alfamovirus of the family Bromoviridae. In 1931 Weimer J.L. was the first to report AMV in alfalfa. Transmission of the virus occurs mainly by some aphids, by seeds or by pollen to the seed.

A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea. Since Dmitri Ivanovsky's 1892 article describing a non-bacterial pathogen infecting tobacco plants and the discovery of the tobacco mosaic virus by Martinus Beijerinck in 1898, more than 9,000 virus species have been described in detail of the millions of types of viruses in the environment. Viruses are found in almost every ecosystem on Earth and are the most numerous type of biological entity. The study of viruses is known as virology, a subspeciality of microbiology.
Vesivirus is a genus of viruses, in the family Caliciviridae. Swine, sea mammals, and felines serve as natural hosts. There are two species in this genus. Diseases associated with this genus include: respiratory disease, Feline calicivirus (FCV); conjunctivitis, and respiratory disease.
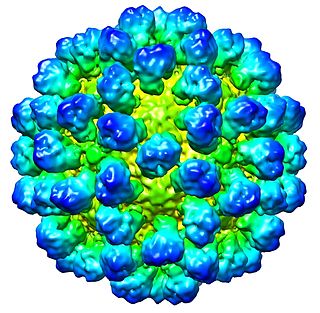
Lagovirus is a genus of viruses, in the family Caliciviridae. Lagomorphs serve as natural hosts. There are two species in this genus. Diseases associated with this genus include: necrotizing hepatitis leading to fatal hemorrhages.
The transmission of hepadnaviruses between their natural hosts, humans, non-human primates, and birds, including intra-species host transmission and cross-species transmission, is a topic of study in virology.
References
- 1 2 3 4 Rowson, K. E. K. (1981). A Dictionary of Virology. Oxford: Blackwell Scientific Publications. ISBN 0632007842.
- ↑ Mahy, Brian W. J. (2001). A Dictionary of Virology (3rd ed.). San Diego, CA: Academic Press. ISBN 9780080546322.
- 1 2 Singleton, Paul; Sainsbury, Diana (2001). Dictionary of Microbiology and Molecular Biology (3rd ed.). Chichester: Wiley. ISBN 0471490644.
