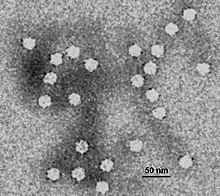
Parvoviruses are a family of animal viruses that constitute the family Parvoviridae. They have linear, single-stranded DNA (ssDNA) genomes that typically contain two genes encoding for a replication initiator protein, called NS1, and the protein the viral capsid is made of. The coding portion of the genome is flanked by telomeres at each end that form into hairpin loops that are important during replication. Parvovirus virions are small compared to most viruses, at 23–28 nanometers in diameter, and contain the genome enclosed in an icosahedral capsid that has a rugged surface.

Carnivore protoparvovirus 1 is a species of parvovirus that infects carnivorans. It causes a highly contagious disease in both dogs and cats separately. The disease is generally divided into two major genogroups: CPV-1 containing the classical feline panleukopenia virus (FPLV), and CPV-2 containing the canine parvovirus (CPV) which appeared in the 1970s.

Canine parvovirus is a contagious virus mainly affecting dogs. CPV is highly contagious and is spread from dog to dog by direct or indirect contact with their feces. Vaccines can prevent this infection, but mortality can reach 91% in untreated cases. Treatment often involves veterinary hospitalization. Canine parvovirus often infects other mammals including foxes, wolves, cats, and skunks. Felines (cats) are also susceptible to panleukopenia, a different strain of parvovirus.
Amdoparvovirus is a genus of viruses in the family Parvoviridae in the subfamily Parvovirinae. Mustelids, skunk, and raccoons serve as natural hosts. There are five species in this genus. Diseases associated with this genus include progressive disorder of immune system.
Carnivore bocaparvovirus 1, formerly Canine minute virus is a species of Bocaparvovirus of the family Parvoviridae that infects dogs. It is similar to bovine parvovirus in its protein structure and DNA. A virus causing respiratory disease in humans has been called human bocavirus due to its similarity to these viruses. Canine minute virus was originally discovered in Germany in 1967 in military dogs, although it was originally thought to not cause disease. Dogs and puppies are infected orally, and the virus is spread transplacentally to the fetuses. Symptoms are seen most commonly between the ages of one to three weeks and include severe diarrhea, difficulty breathing, and anorexia. In severe cases, illness can be fatal.

Dependoparvovirus is a genus in the subfamily Parvovirinae of the virus family Parvoviridae; they are Group II viruses according to the Baltimore classification. Some dependoparvoviruses are also known as adeno-associated viruses because they cannot replicate productively in their host cell without the cell being coinfected by a helper virus such as an adenovirus, a herpesvirus, or a vaccinia virus.
Human bocavirus (HBoV) is the name given to all viruses in the genus Bocaparvovirus of virus family Parvoviridae that are known to infect humans. HBoV1 and HBoV3 are members of species Primate bocaparvovirus 1 whereas viruses HBoV2 and HBoV4 belong to species Primate bocaparvovirus 2. Some of these viruses cause human disease. HBoV1 is strongly implicated in causing some cases of lower respiratory tract infection, especially in young children, and several of the viruses have been linked to gastroenteritis, although the full clinical role of this emerging infectious disease remains to be elucidated.

Erythroparvovirus is a genus of viruses in subfamily Parvovirinae of the virus family Parvoviridae. Primates serve as natural hosts. There are seven species in this genus. Diseases associated with this genus include fifth disease and skin lesions.
Mink enteritis virus (MEV) is a strain of Carnivore protoparvovirus 1 that infects mink and causes enteritis. Like all parvoviruses, it is a small, spherical virus, and has a single-stranded DNA genome. The signs and symptoms of enteritis usually appear within 4–7 days after infection. The virus replicates in the cells of the crypt epithelium in the duodenum and jejunum and, to a lesser extent the ileum, colon and caecum. The severity of the disease is directly related to necrosis of the crypt epithelium.
Bocaparvovirus is a genus of viruses in the subfamily Parvovirinae of the virus family Parvoviridae. Humans, cattle, and dogs serve as natural hosts. There are 28 species in this genus. Diseases associated with this genus include, in humans, acute respiratory illness, and in cattle, diarrhea and mild respiratory symptoms.
Densovirinae is a subfamily of single-stranded DNA viruses in the family Parvoviridae. The subfamily has 11 recognized genera and 21 species. Densoviruses are known to infect members of insect orders Blattodea, Diptera, Hemiptera, Hymenoptera, Lepidoptera, and Orthoptera, while some viruses infect and multiply in crustaceans such as shrimp or crayfish, or sea stars from phylum Echinodermata.
Tetraparvovirus are a genus of viruses in the family Parvoviridae. There are six recognized species: Chiropteran tetraparvovirus 1, Primate tetraparvovirus 1, Ungulate tetraparvovirus 1, Ungulate tetraparvovirus 2, Ungulate tetraparvovirus 3, and Ungulate tetraparvovirus 4.

Minute virus of mice (MVM) is the exemplar virus of the species Rodent protoparvovirus 1, in the genus Protoparvovirus of the Parvoviridae family of viruses. MVM exists in multiple variant forms including MVMp, which is the prototype strain that infects cells of fibroblast origin, while MVMi, the immunosuppressive strain, infects T lymphocytes. MVM is a common infection in laboratory mice due to its highly contagious nature. The virus can be shed from infected mice via feces and urine, but also via fomites and nasal secretions. Typically there are no clinical signs of infection in adult mice, however, experimental infection can cause multiple organ damage during fetal development or shortly after birth.
Brevihamaparvovirus is a genus of viruses in subfamily Hamaparvovirinae of the family Parvoviridae. Mosquitoes serve as natural hosts. There are two species in this genus.
Iteradensovirus is a genus of viruses in the subfamily Densovirinae of the family Parvoviridae. Insects serve as natural hosts. There are five species in this genus.
Aveparvovirus is a genus of viruses, in the subfamily Parvovirinae of the virus family Parvoviridae. There are three species in this genus. Diseases associated with this genus include: enteric disease and malabsorption syndrome.
Copiparvovirus is a genus of viruses in subfamily Parvovirinae of the virus family Parvoviridae. Pigs and cows are known to serve as natural hosts. There are seven species in this genus.
Hepanhamaparvovirus is a genus of viruses that belongs to the Hapanhamavirinae subfamily of the family Parvoviridae. Insects and shrimps serve as natural hosts. Infection leads to mortality in the early larval and postlarval stages of the shrimp. There is only one species in this genus: Decapod hepanhamaparvovirus 1.
Penstylhamaparvovirus is the name of a genus of viruses in the subfamily Hamaparvovirinae of the virus family Parvoviridae. Shrimps and insects serve as natural hosts. There is only one species in this genus: Decapod penstylhamaparvovirus 1.
Rolling hairpin replication (RHR) is a unidirectional, strand displacement form of DNA replication used by parvoviruses, a group of viruses that constitute the family Parvoviridae. Parvoviruses have linear, single-stranded DNA (ssDNA) genomes in which the coding portion of the genome is flanked by telomeres at each end that form hairpin loops. During RHR, these hairpin loops repeatedly unfold and refold to change the direction of DNA replication so that replication progresses in a continuous manner back and forth across the genome. RHR is initiated and terminated by an endonuclease encoded by parvoviruses that is variously called NS1 or Rep, and RHR is similar to rolling circle replication, which is used by ssDNA viruses that have circular genomes.