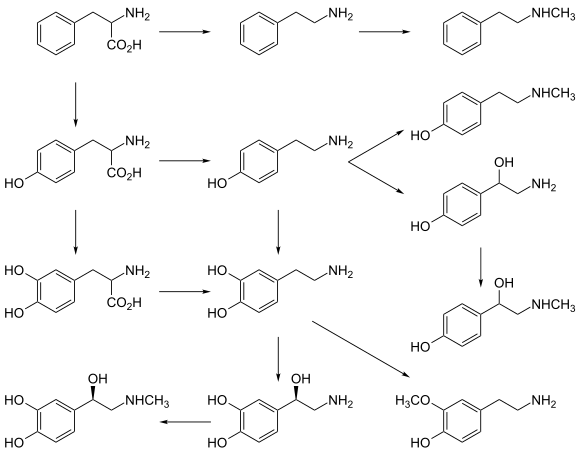
A catecholamine is a monoamine neurotransmitter, an organic compound that has a catechol and a side-chain amine.

Monoamine neurotransmitters are neurotransmitters and neuromodulators that contain one amino group connected to an aromatic ring by a two-carbon chain (such as -CH2-CH2-). Examples are dopamine, norepinephrine and serotonin.

Phenethylamine (PEA) is an organic compound, natural monoamine alkaloid, and trace amine, which acts as a central nervous system stimulant in humans. In the brain, phenethylamine regulates monoamine neurotransmission by binding to trace amine-associated receptor 1 (TAAR1) and inhibiting vesicular monoamine transporter 2 (VMAT2) in monoamine neurons. To a lesser extent, it also acts as a neurotransmitter in the human central nervous system. In mammals, phenethylamine is produced from the amino acid L-phenylalanine by the enzyme aromatic L-amino acid decarboxylase via enzymatic decarboxylation. In addition to its presence in mammals, phenethylamine is found in many other organisms and foods, such as chocolate, especially after microbial fermentation.
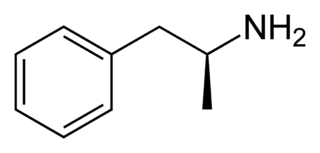
Dextroamphetamine (INN:dexamfetamine) is a potent central nervous system (CNS) stimulant and enantiomer of amphetamine that is prescribed for the treatment of attention deficit hyperactivity disorder (ADHD) and narcolepsy. It is also used as an athletic performance and cognitive enhancer, and recreationally as an aphrodisiac and euphoriant. Dextroamphetamine is generally regarded as the prototypical stimulant.

Phenylpropanolamine (PPA), sold under many brand names, is a sympathomimetic agent which is used as a decongestant and appetite suppressant. It was previously commonly used in prescription and over-the-counter cough and cold preparations. The medication is taken by mouth.

The norepinephrine transporter (NET), also known as noradrenaline transporter (NAT), is a protein that in humans is encoded by the solute carrier family 6 member 2 (SLC6A2) gene.

Phenylacetone, also known as phenyl-2-propanone, is an organic compound with the chemical formula C6H5CH2COCH3. It is a colorless oil that is soluble in organic solvents. It is a mono-substituted benzene derivative, consisting of an acetone attached to a phenyl group. As such, its systematic IUPAC name is 1-phenyl-2-propanone.
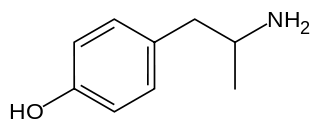
Hydroxyamphetamine, also known as 4-hydroxyamphetamine or norpholedrine and sold under the brand names Paredrine and Paremyd among others, is a sympathomimetic medication used in eye drops to dilate the pupil for eye examinations.
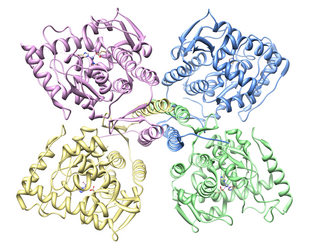
Tyrosine hydroxylase or tyrosine 3-monooxygenase is the enzyme responsible for catalyzing the conversion of the amino acid L-tyrosine to L-3,4-dihydroxyphenylalanine (L-DOPA). It does so using molecular oxygen (O2), as well as iron (Fe2+) and tetrahydrobiopterin as cofactors. L-DOPA is a precursor for dopamine, which, in turn, is a precursor for the important neurotransmitters norepinephrine (noradrenaline) and epinephrine (adrenaline). Tyrosine hydroxylase catalyzes the rate limiting step in this synthesis of catecholamines. In humans, tyrosine hydroxylase is encoded by the TH gene, and the enzyme is present in the central nervous system (CNS), peripheral sympathetic neurons and the adrenal medulla. Tyrosine hydroxylase, phenylalanine hydroxylase and tryptophan hydroxylase together make up the family of aromatic amino acid hydroxylases (AAAHs).

Steroid 21-hydroxylase is a protein that in humans is encoded by the CYP21A2 gene. The protein is an enzyme that hydroxylates steroids at the C21 position on the molecule. Naming conventions for enzymes are based on the substrate acted upon and the chemical process performed. Biochemically, this enzyme is involved in the biosynthesis of the adrenal gland hormones aldosterone and cortisol, which are important in blood pressure regulation, sodium homeostasis and blood sugar control. The enzyme converts progesterone and 17α-hydroxyprogesterone into 11-deoxycorticosterone and 11-deoxycortisol, respectively, within metabolic pathways which in humans ultimately lead to aldosterone and cortisol creation—deficiency in the enzyme may cause congenital adrenal hyperplasia.

Steroid 11β-hydroxylase, also known as steroid 11β-monooxygenase, is a steroid hydroxylase found in the zona glomerulosa and zona fasciculata of the adrenal cortex. Named officially the cytochrome P450 11B1, mitochondrial, it is a protein that in humans is encoded by the CYP11B1 gene. The enzyme is involved in the biosynthesis of adrenal corticosteroids by catalyzing the addition of hydroxyl groups during oxidation reactions.

Lisdexamfetamine, sold under the brand names Vyvanse and Elvanse among others, is a stimulant medication that is used to treat attention deficit hyperactivity disorder (ADHD) in children and adults and for moderate-to-severe binge eating disorder in adults. Lisdexamfetamine is taken by mouth. Its effects generally begin within two hours and last for up to 14 hours.

In enzymology, a kynurenine 3-monooxygenase (EC 1.14.13.9) is an enzyme that catalyzes the chemical reaction

Nepicastat is an inhibitor of dopamine beta-hydroxylase (DBH), an enzyme that catalyzes the conversion of dopamine to norepinephrine.

Dopamine beta (β)-hydroxylase deficiency is a human medical condition involving inadequate dopamine beta-hydroxylase. It is characterized by increased amounts of serum dopamine and the absence of norepinephrine (NE) and epinephrine.

3,4-Dihydroxyphenylacetaldehyde (DOPAL), also known as dopamine aldehyde, is a metabolite of the monoamine neurotransmitter dopamine formed by monoamine oxidase (MAO).

DBH-like monooxygenase protein 1, also known as monooxygenase X, is an enzyme that in humans is encoded by the MOXD1 gene.

p-Hydroxynorephedrine is the para-hydroxy analog of norephedrine and an active sympathomimetic metabolite of amphetamine in humans. When it occurs as a metabolite of amphetamine, it is produced from both p-hydroxyamphetamine and norephedrine.

4-Hydroxyphenylacetone is the para-hydroxy analog of phenylacetone, an inactive metabolite of amphetamine in humans. When it occurs as a metabolite of amphetamine, it is produced directly from the inactive metabolite phenylacetone.
Cytochrome P450 omega hydroxylases, also termed cytochrome P450 ω-hydroxylases, CYP450 omega hydroxylases, CYP450 ω-hydroxylases, CYP omega hydroxylase, CYP ω-hydroxylases, fatty acid omega hydroxylases, cytochrome P450 monooxygenases, and fatty acid monooxygenases, are a set of cytochrome P450-containing enzymes that catalyze the addition of a hydroxyl residue to a fatty acid substrate. The CYP omega hydroxylases are often referred to as monoxygenases; however, the monooxygenases are CYP450 enzymes that add a hydroxyl group to a wide range of xenobiotic and naturally occurring endobiotic substrates, most of which are not fatty acids. The CYP450 omega hydroxylases are accordingly better viewed as a subset of monooxygenases that have the ability to hydroxylate fatty acids. While once regarded as functioning mainly in the catabolism of dietary fatty acids, the omega oxygenases are now considered critical in the production or break-down of fatty acid-derived mediators which are made by cells and act within their cells of origin as autocrine signaling agents or on nearby cells as paracrine signaling agents to regulate various functions such as blood pressure control and inflammation.