
A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism, and cell signaling.
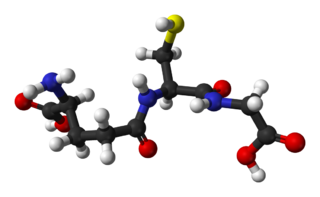
Glutathione is an antioxidant in plants, animals, fungi, and some bacteria and archaea. Glutathione is capable of preventing damage to important cellular components caused by sources such as reactive oxygen species, free radicals, peroxides, lipid peroxides, and heavy metals. It is a tripeptide with a gamma peptide linkage between the carboxyl group of the glutamate side chain and cysteine. The carboxyl group of the cysteine residue is attached by normal peptide linkage to glycine.

In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the binding site, and residues that catalyse a reaction of that substrate, the catalytic site. Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes.

Antithrombin (AT) is a small glycoprotein that inactivates several enzymes of the coagulation system. It is a 464-amino-acid protein produced by the liver. It contains three disulfide bonds and a total of four possible glycosylation sites. α-Antithrombin is the dominant form of antithrombin found in blood plasma and has an oligosaccharide occupying each of its four glycosylation sites. A single glycosylation site remains consistently un-occupied in the minor form of antithrombin, β-antithrombin. Its activity is increased manyfold by the anticoagulant drug heparin, which enhances the binding of antithrombin to factor IIa (thrombin) and factor Xa.

α2-Macroglobulin (α2M), or alpha-2-macroglobulin, is a large plasma protein found in the blood. It is mainly produced by the liver, and also locally synthesized by macrophages, fibroblasts, and adrenocortical cells. In humans it is encoded by the A2M gene.

Serine proteases are enzymes that cleave peptide bonds in proteins. Serine serves as the nucleophilic amino acid at the (enzyme's) active site. They are found ubiquitously in both eukaryotes and prokaryotes. Serine proteases fall into two broad categories based on their structure: chymotrypsin-like (trypsin-like) or subtilisin-like.

Tienilic acid or ticrynafen (USAN) is a loop diuretic drug with uric acid-lowering (uricosuric) action, formerly marketed for the treatment of hypertension. It was approved by FDA on May 2, 1979, and withdrawn in 1982, after case reports in the United States indicated a link between the use of ticrynafen and hepatitis.

A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes. An acid-base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence.

Dithiothreitol (DTT) is the common name for a small-molecule redox reagent also known as Cleland's reagent, after W. Wallace Cleland. DTT's formula is C4H10O2S2 and the chemical structure of one of its enantiomers in its reduced form is shown on the right; its oxidized form is a disulfide bonded 6-membered ring (shown below). The reagent is commonly used in its racemic form, as both enantiomers are reactive. Its name derives from the four-carbon sugar, threose. DTT has an epimeric ('sister') compound, dithioerythritol (DTE).

2-Iodoacetamide is an alkylating agent used for peptide mapping purposes. Its actions are similar to those of iodoacetate. It is commonly used to bind covalently with the thiol group of cysteine so the protein cannot form disulfide bonds. Also used in ubiquitin studies as an inhibitor of deubiquitinase enzymes (DUBs) because it alkylates the cysteine residues at the DUB active site.

Cysteine proteases, also known as thiol proteases, are hydrolase enzymes that degrade proteins. These proteases share a common catalytic mechanism that involves a nucleophilic cysteine thiol in a catalytic triad or dyad.
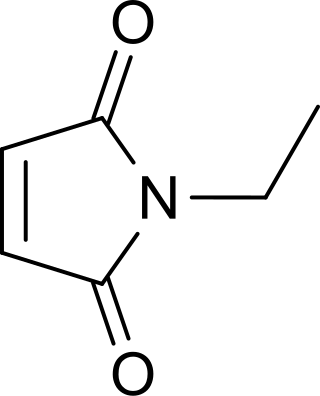
N-Ethylmaleimide (NEM) is an organic compound that is derived from maleic acid. It contains the amide functional group, but more importantly it is an alkene that is reactive toward thiols and is commonly used to modify cysteine residues in proteins and peptides.
A carboxypeptidase is a protease enzyme that hydrolyzes (cleaves) a peptide bond at the carboxy-terminal (C-terminal) end of a protein or peptide. This is in contrast to an aminopeptidases, which cleave peptide bonds at the N-terminus of proteins. Humans, animals, bacteria and plants contain several types of carboxypeptidases that have diverse functions ranging from catabolism to protein maturation. At least two mechanisms have been discussed.
Protein metabolism denotes the various biochemical processes responsible for the synthesis of proteins and amino acids (anabolism), and the breakdown of proteins by catabolism.

Actinidain is a type of cysteine protease enzyme found in fruits including kiwifruit, pineapple, mango, banana, figs, and papaya. This enzyme is part of the peptidase C1 family of papain-like proteases.
MEROPS is an online database for peptidases and their inhibitors. The classification scheme for peptidases was published by Rawlings & Barrett in 1993, and that for protein inhibitors by Rawlings et al. in 2004. The most recent version, MEROPS 12.4, was released in late October 2021.

In molecular biology Proteinase K is a broad-spectrum serine protease. The enzyme was discovered in 1974 in extracts of the fungus Engyodontium album. Proteinase K is able to digest hair (keratin), hence, the name "Proteinase K". The predominant site of cleavage is the peptide bond adjacent to the carboxyl group of aliphatic and aromatic amino acids with blocked alpha amino groups. It is commonly used for its broad specificity. This enzyme belongs to Peptidase family S8 (subtilisin). The molecular weight of Proteinase K is 28,900 daltons.

An Oligopeptidase is an enzyme that cleaves peptides but not proteins. This property is due to its structure: the active site of this enzyme is located at the end of a narrow cavity which can only be reached by peptides.

Zingibain, zingipain, or ginger protease is a cysteine protease enzyme found in ginger rhizomes. It catalyses the preferential cleavage of peptides with a proline residue at the P2 position. It has two distinct forms, ginger protease I (GP-I) and ginger protease II (GP-II).

4-Aminophenylmercuric acetate (CH3CO2HgC6H4NH2, also known as 4-(Acetoxymercurio)aniline or APMA), is an organomercurial compound and thiol-blocking reagent used in experimental biology and chemistry to activate matrix metalloproteinases and collagenase proteolytic enzymes. The material is highly toxic.






















