
A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.

Ribonuclease is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 and 3.1 classes of enzymes.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.

Ribosomal DNA (rDNA) is a DNA sequence that codes for ribosomal RNA. These sequences regulate transcription initiation and amplification, and contain both transcribed and non-transcribed spacer segments.

Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass.

Cartilage–hair hypoplasia (CHH) is a rare genetic disorder. Symptoms may include short-limbed dwarfism due to skeletal dysplasia, variable level of immunodeficiency, and predisposition to cancer. It was first reported by Victor McKusick in 1965.
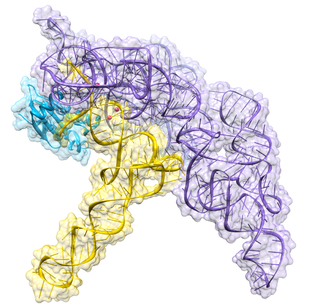
Ribonuclease P is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein-based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules. Further, RNase P is one of two known multiple turnover ribozymes in nature, the discovery of which earned Sidney Altman and Thomas Cech the Nobel Prize in Chemistry in 1989: in the 1970s, Altman discovered the existence of precursor tRNA with flanking sequences and was the first to characterize RNase P and its activity in processing of the 5' leader sequence of precursor tRNA. Recent findings also reveal that RNase P has a new function. It has been shown that human nuclear RNase P is required for the normal and efficient transcription of various small noncoding RNAs, such as tRNA, 5S rRNA, SRP RNA and U6 snRNA genes, which are transcribed by RNA polymerase III, one of three major nuclear RNA polymerases in human cells.

The exosome complex is a multi-protein intracellular complex capable of degrading various types of RNA molecules. Exosome complexes are found in both eukaryotic cells and archaea, while in bacteria a simpler complex called the degradosome carries out similar functions.

Eukaryotic transcription is the elaborate process that eukaryotic cells use to copy genetic information stored in DNA into units of transportable complementary RNA replica. Gene transcription occurs in both eukaryotic and prokaryotic cells. Unlike prokaryotic RNA polymerase that initiates the transcription of all different types of RNA, RNA polymerase in eukaryotes comes in three variations, each translating a different type of gene. A eukaryotic cell has a nucleus that separates the processes of transcription and translation. Eukaryotic transcription occurs within the nucleus where DNA is packaged into nucleosomes and higher order chromatin structures. The complexity of the eukaryotic genome necessitates a great variety and complexity of gene expression control.
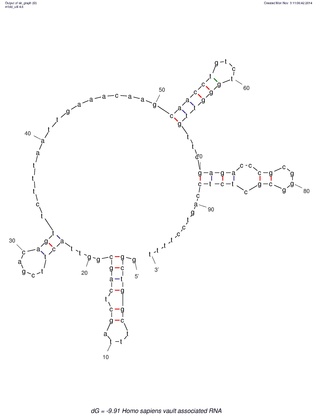
Many eukaryotic cells contain large ribonucleoprotein particles in the cytoplasm known as vaults. The vault complex comprises the major vault protein (MVP), two minor vault proteins, and a variety of small untranslated RNA molecules known as vault RNAs only found in higher eukaryotes. These molecules are transcribed by RNA polymerase III.
Trans-regulatory elements (TRE) are DNA sequences encoding upstream regulators, which may modify or regulate the expression of distant genes. Trans-acting factors interact with cis-regulatory elements to regulate gene expression. TRE mediates expression profiles of a large number of genes via trans-acting factors. While TRE mutations affect gene expression, it is also one of the main driving factors for evolutionary divergence in gene expression.

Ribonucleases P/MRP protein subunit POP1 is a protein that in humans is encoded by the POP1 gene.

Ribonuclease P protein subunit p30 is an enzyme that in humans is encoded by the RPP30 gene.

Ribonuclease P protein subunit p38 is an enzyme that in humans is encoded by the RPP38 gene.

Ribonuclease P protein subunit p14 is an enzyme that in humans is encoded by the RPP14 gene.

RNA component of mitochondrial RNA processing endoribonuclease, also known as RMRP, is a human gene.

Ribonuclease 4 is an enzyme that in humans is encoded by the RNASE4 gene.

The 5′ flanking region is a region of DNA that is adjacent to the 5′ end of the gene. The 5′ flanking region contains the promoter, and may contain enhancers or other protein binding sites. It is the region of DNA that is not transcribed into RNA. Not to be confused with the 5′ untranslated region, this region is not transcribed into RNA or translated into a functional protein. These regions primarily function in the regulation of gene transcription. 5′ flanking regions are categorized between prokaryotes and eukaryotes.

ATP-binding cassette sub-family E member 1 (ABCE1) also known as RNase L inhibitor (RLI) is an enzyme that in humans is encoded by the ABCE1 gene.

ZC3H12B, also known as CXorf32 or MCPIP2, is a protein encoded by gene ZC3H12B located on chromosome Xq12 in humans.





















