
Agarose gel electrophoresis is a method of gel electrophoresis used in biochemistry, molecular biology, genetics, and clinical chemistry to separate a mixed population of macromolecules such as DNA or proteins in a matrix of agarose, one of the two main components of agar. The proteins may be separated by charge and/or size, and the DNA and RNA fragments by length. Biomolecules are separated by applying an electric field to move the charged molecules through an agarose matrix, and the biomolecules are separated by size in the agarose gel matrix.

Gel electrophoresis is a method for separation and analysis of biomacromolecules and their fragments, based on their size and charge. It is used in clinical chemistry to separate proteins by charge or size and in biochemistry and molecular biology to separate a mixed population of DNA and RNA fragments by length, to estimate the size of DNA and RNA fragments or to separate proteins by charge.
The isoelectric point (pI, pH(I), IEP), is the pH at which a molecule carries no net electrical charge or is electrically neutral in the statistical mean. The standard nomenclature to represent the isoelectric point is pH(I). However, pI is also used. For brevity, this article uses pI. The net charge on the molecule is affected by pH of its surrounding environment and can become more positively or negatively charged due to the gain or loss, respectively, of protons (H+).

Southern blot is a method used for detection and quantification of a specific DNA sequence in DNA samples. This method is used in molecular biology. Briefly, purified DNA from a biological sample is digested with restriction enzymes, and the resulting DNA fragments are separated by using an electric current to move them through a sieve-like gel or matrix, which allows smaller fragments to move faster than larger fragments. The DNA fragments are transferred out of the gel or matrix onto a solid membrane, which is then exposed to a DNA probe labeled with a radioactive, fluorescent, or chemical tag. The tag allows any DNA fragments containing complementary sequences with the DNA probe sequence to be visualized within the Southern blot.
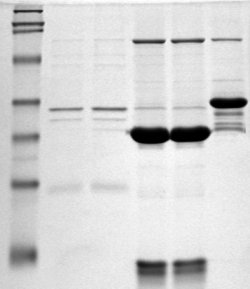
Polyacrylamide gel electrophoresis (PAGE) is a technique widely used in biochemistry, forensic chemistry, genetics, molecular biology and biotechnology to separate biological macromolecules, usually proteins or nucleic acids, according to their electrophoretic mobility. Electrophoretic mobility is a function of the length, conformation, and charge of the molecule. Polyacrylamide gel electrophoresis is a powerful tool used to analyze RNA samples. When polyacrylamide gel is denatured after electrophoresis, it provides information on the sample composition of the RNA species.

Two-dimensional gel electrophoresis, abbreviated as 2-DE or 2-D electrophoresis, is a form of gel electrophoresis commonly used to analyze proteins. Mixtures of proteins are separated by two properties in two dimensions on 2D gels. 2-DE was first independently introduced by O'Farrell and Klose in 1975.

Protein electrophoresis is a method for analysing the proteins in a fluid or an extract. The electrophoresis may be performed with a small volume of sample in a number of alternative ways with or without a supporting medium, namely agarose or polyacrylamide. Variants of gel electrophoresis include SDS-PAGE, free-flow electrophoresis, electrofocusing, isotachophoresis, affinity electrophoresis, immunoelectrophoresis, counterelectrophoresis, and capillary electrophoresis. Each variant has many subtypes with individual advantages and limitations. Gel electrophoresis is often performed in combination with electroblotting or immunoblotting to give additional information about a specific protein.
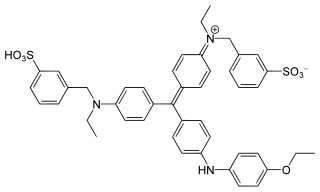
Coomassie brilliant blue is the name of two similar triphenylmethane dyes that were developed for use in the textile industry but are now commonly used for staining proteins in analytical biochemistry. Coomassie brilliant blue G-250 differs from Coomassie brilliant blue R-250 by the addition of two methyl groups. The name "Coomassie" is a registered trademark of Imperial Chemical Industries.

Isoelectric focusing (IEF), also known as electrofocusing, is a technique for separating different molecules by differences in their isoelectric point (pI). It is a type of zone electrophoresis usually performed on proteins in a gel that takes advantage of the fact that overall charge on the molecule of interest is a function of the pH of its surroundings.
In pathology, silver staining is the use of silver to selectively alter the appearance of a target in microscopy of histological sections; in temperature gradient gel electrophoresis; and in polyacrylamide gels.

MOPS (3-(N-morpholino)propanesulfonic acid) is a buffer introduced in the 1960s, one of the twenty Good's buffers. It is a structural analog to MES, and like MES, its structure contains a morpholine ring. HEPES is a similar pH buffering compound that contains a piperazine ring. With a pKa of 7.20, MOPS is an excellent buffer for many biological systems at near-neutral pH.

Tricine is an organic compound that is used in buffer solutions. The name tricine comes from tris and glycine, from which it was derived. It is a white crystalline powder that is moderately soluble in water. It is a zwitterionic amino acid that has a pKa1 value of 2.3 at 25 °C, while its pKa2 at 20 °C is 8.15. Its useful buffering range of pH is 7.4-8.8. Along with bicine, it is one of Good's buffering agents. Good first prepared tricine to buffer chloroplast reactions.

A molecular-weight size marker, also referred to as a protein ladder, DNA ladder, or RNA ladder, is a set of standards that are used to identify the approximate size of a molecule run on a gel during electrophoresis, using the principle that molecular weight is inversely proportional to migration rate through a gel matrix. Therefore, when used in gel electrophoresis, markers effectively provide a logarithmic scale by which to estimate the size of the other fragments.
QPNC-PAGE, or Quantitative Preparative Native Continuous PolyAcrylamide Gel Electrophoresis, is a bioanalytical, one-dimensional, high-resolution and high-precision technique applied in biochemistry and bioinorganic chemistry to separate proteins quantitatively by isoelectric point and by continuous elution from a gel column. This standardized variant of native gel electrophoresis and subset of preparative polyacrylamide gel electrophoresis is used by biologists to isolate macromolecules in solution, for example, active or native metalloproteins in biological samples or properly and improperly folded metal cofactor-containing proteins or protein isoforms in complex protein mixtures.
Within chemistry for acid–base reactions, Immobilized pH gradient (IPG) gels are the acrylamide gel matrix co-polymerized with the pH gradient, which result in completely stable gradients except the most alkaline (>12) pH values. The immobilized pH gradient is obtained by the continuous change in the ratio of Immobilines. An Immobiline is a weak acid or base defined by its pK value. Immobilized pH gradients (IPG) are made by mixing two kinds of acrylamide mixture, one with Immobiline having acidic buffering property and other with basic buffering property. The concentrations of the buffers in the two solutions define the range and shape of the pH gradient produced. Both solutions contain acrylamide monomers and catalysts. During polymerization, the acrylamide portion of the buffers co polymerize with the acrylamide and bisacrylamide monomers to form a polyacrylamide gel. These polymerised gels are backed with plastic based backing that allow ease in handling and improve IPG's performance. The gel is then washed to remove catalysts and unpolymerized monomers, which interfere with isoelectric separation. IPG increased reproducibility of Isoelectric focusing and 2D-Gel Electrophoresis. Other advantages are increased resolution, reproducible separation of alkaline proteins and increased loading capacity.

An electrophoretic color marker is a chemical used to monitor the progress of agarose gel electrophoresis and polyacrylamide gel electrophoresis (PAGE) since DNA, RNA, and most proteins are colourless. The color markers are made up of a mixture of dyes that migrate through the gel matrix alongside the sample of interest. They are typically designed to have different mobilities from the sample components and to generate colored bands that can be used to assess the migration and separation of sample components.

Affinity electrophoresis is a general name for many analytical methods used in biochemistry and biotechnology. Both qualitative and quantitative information may be obtained through affinity electrophoresis. Cross electrophoresis, the first affinity electrophoresis method, was created by Nakamura et al. Enzyme-substrate complexes have been detected using cross electrophoresis. The methods include the so-called electrophoretic mobility shift assay, charge shift electrophoresis and affinity capillary electrophoresis. The methods are based on changes in the electrophoretic pattern of molecules through biospecific interaction or complex formation. The interaction or binding of a molecule, charged or uncharged, will normally change the electrophoretic properties of a molecule. Membrane proteins may be identified by a shift in mobility induced by a charged detergent. Nucleic acids or nucleic acid fragments may be characterized by their affinity to other molecules. The methods have been used for estimation of binding constants, as for instance in lectin affinity electrophoresis or characterization of molecules with specific features like glycan content or ligand binding. For enzymes and other ligand-binding proteins, one-dimensional electrophoresis similar to counter electrophoresis or to "rocket immunoelectrophoresis", affinity electrophoresis may be used as an alternative quantification of the protein. Some of the methods are similar to affinity chromatography by use of immobilized ligands.

MOPSO is a zwitterionic organic chemical buffering agent; one of Good's buffers. MOPSO and MOPS are chemically similar, differing only in the presence of a hydroxyl group on the C-2 of the propane moiety. It has a useful pH range of 6.5-7.9 in the physiological range, making it useful for cell culture work. It has a pKa of 6.9 with ΔpKa/°C of -0.015 and a solubility in water at 0°C of 0.75 M.
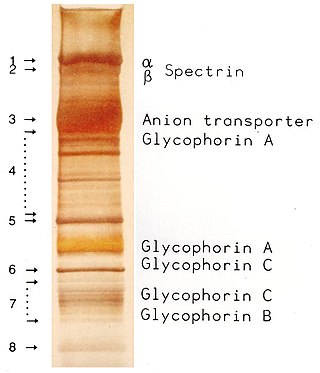
SDS-PAGE is a discontinuous electrophoretic system developed by Ulrich K. Laemmli which is commonly used as a method to separate proteins with molecular masses between 5 and 250 kDa. The combined use of sodium dodecyl sulfate and polyacrylamide gel eliminates the influence of structure and charge, and proteins are separated by differences in their size. At least up to 2012, the publication describing it was the most frequently cited paper by a single author, and the second most cited overall.

N,N′-Diallyl-L-tartardiamide (DATD) is a crosslinking agent for polyacrylamide gels, e.g., as used for SDS-PAGE. Compared to bisacrylamide gels, DATD gels have a stronger interaction with glass, and therefore are used in applications where the polyacrylamide gel acts as a "plug" structural component at the bottom of a gel electrophoresis apparatus, thereby preventing a weak discontinuous gel from sliding out from or otherwise moving within the apparatus. Unlike bisacrylamide-polyacrylamide gels, DATD-polyacrylamide gels can be conveniently dissolved using periodic acid due to the presence of viscinal diols in DATD. DATD is the slowest polyacrylamide crosslinker tested, and has can act as an inhibitor of polymerization at high concentrations.















