Computational biology refers to the use of data analysis, mathematical modeling and computational simulations to understand biological systems and relationships. An intersection of computer science, biology, and big data, the field also has foundations in applied mathematics, chemistry, and genetics. It differs from biological computing, a subfield of computer science and engineering which uses bioengineering to build computers.
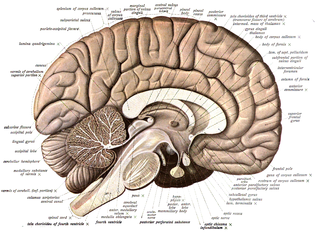
Neuroanatomy is the study of the structure and organization of the nervous system. In contrast to animals with radial symmetry, whose nervous system consists of a distributed network of cells, animals with bilateral symmetry have segregated, defined nervous systems. Their neuroanatomy is therefore better understood. In vertebrates, the nervous system is segregated into the internal structure of the brain and spinal cord and the series of nerves that connect the CNS to the rest of the body. Breaking down and identifying specific parts of the nervous system has been crucial for figuring out how it operates. For example, much of what neuroscientists have learned comes from observing how damage or "lesions" to specific brain areas affects behavior or other neural functions.

A neuroscientist is a scientist who has specialised knowledge in neuroscience, a branch of biology that deals with the physiology, biochemistry, psychology, anatomy and molecular biology of neurons, neural circuits, and glial cells and especially their behavioral, biological, and psychological aspect in health and disease.
Neuroinformatics is the field that combines informatics and neuroscience. Neuroinformatics is related with neuroscience data and information processing by artificial neural networks. There are three main directions where neuroinformatics has to be applied:
Brain mapping is a set of neuroscience techniques predicated on the mapping of (biological) quantities or properties onto spatial representations of the brain resulting in maps.

The Allen Institute for Brain Science is a division of the Allen Institute, based in Seattle, Washington, that focuses on bioscience research. Founded in 2003, it is dedicated to accelerating the understanding of how the human brain works. With the intent of catalyzing brain research in different areas, the Allen Institute provides free data and tools to scientists.

Neurogenetics studies the role of genetics in the development and function of the nervous system. It considers neural characteristics as phenotypes, and is mainly based on the observation that the nervous systems of individuals, even of those belonging to the same species, may not be identical. As the name implies, it draws aspects from both the studies of neuroscience and genetics, focusing in particular how the genetic code an organism carries affects its expressed traits. Mutations in this genetic sequence can have a wide range of effects on the quality of life of the individual. Neurological diseases, behavior and personality are all studied in the context of neurogenetics. The field of neurogenetics emerged in the mid to late 20th century with advances closely following advancements made in available technology. Currently, neurogenetics is the center of much research utilizing cutting edge techniques.
Connectomics is the production and study of connectomes: comprehensive maps of connections within an organism's nervous system. More generally, it can be thought of as the study of neuronal wiring diagrams with a focus on how structural connectivity, individual synapses, cellular morphology, and cellular ultrastructure contribute to the make up of a network. The nervous system is a network made of billions of connections and these connections are responsible for our thoughts, emotions, actions, memories, function and dysfunction. Therefore, the study of connectomics aims to advance our understanding of mental health and cognition by understanding how cells in the nervous system are connected and communicate. Because these structures are extremely complex, methods within this field use a high-throughput application of functional and structural neural imaging, most commonly magnetic resonance imaging (MRI), electron microscopy, and histological techniques in order to increase the speed, efficiency, and resolution of these nervous system maps. To date, tens of large scale datasets have been collected spanning the nervous system including the various areas of cortex, cerebellum, the retina, the peripheral nervous system and neuromuscular junctions.
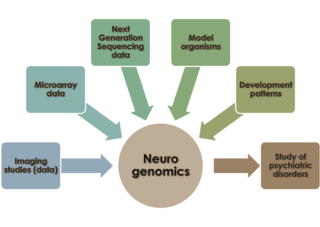
Neurogenomics is the study of how the genome of an organism influences the development and function of its nervous system. This field intends to unite functional genomics and neurobiology in order to understand the nervous system as a whole from a genomic perspective.

Homeobox protein Nkx-6.2 is a protein that in humans is encoded by the NKX6-2 gene.
The Human Connectome Project (HCP) is a five-year project sponsored by sixteen components of the National Institutes of Health, split between two consortia of research institutions. The project was launched in July 2009 as the first of three Grand Challenges of the NIH's Blueprint for Neuroscience Research. On September 15, 2010, the NIH announced that it would award two grants: $30 million over five years to a consortium led by Washington University in St. Louis and the University of Minnesota, with strong contributions from University of Oxford (FMRIB) and $8.5 million over three years to a consortium led by Harvard University, Massachusetts General Hospital and the University of California Los Angeles.
EMAGE is an online biological database of gene expression data in the developing mouse embryo. The data held in EMAGE is spatially annotated to a framework of 3D mouse embryo models produced by EMAP. These spatial annotations allow users to query EMAGE by spatial pattern as well as by gene name, anatomy term or Gene Ontology (GO) term. EMAGE is a freely available web-based resource funded by the Medical Research Council (UK) and based at the MRC Human Genetics Unit in the Institute of Genetics and Molecular Medicine, Edinburgh, UK.

Angela Jane Roskams is a neuroscientist at the University of British Columbia (UBC) with a joint appointment in Neurosurgery at the University of Washington. She is professor at the Centre for Brain Health at UBC, and directed the laboratory of neural regeneration and brain repair, before winding down her lab in 2015–16 to become Executive Director of the Allen Institute for Brain Science, and a leader in the Open Science movement. After leading Strategy and Alliances for the Allen institute's multiple branches, she has become an influencer in the fields of neuroinformatics, public-private partnerships, and Open Data Sharing.
The Human Protein Atlas (HPA) is a Swedish-based program started in 2003 with the aim to map all the human proteins in cells, tissues and organs using integration of various omics technologies, including antibody-based imaging, mass spectrometry-based proteomics, transcriptomics and systems biology. All the data in the knowledge resource is open access to allow scientists both in academia and industry to freely access the data for exploration of the human proteome. In June 2023, version 23 was launched where a new Interaction section was introduced containing human protein-protein interaction networks for more than 11,000 genes that will add new aspects in terms of protein function.
The following outline is provided as an overview of and topical guide to brain mapping:

The Allen Institute is a non-profit, bioscience research institute located in Seattle. It was founded by billionaire philanthropist Paul G. Allen in 2003. The Allen Institute conducts large-scale basic science research studying the brain, cells and immune system in effort to accelerate science and disease research. The organization practices open science, in that they make all their data and resources publicly available for researchers to access.
Jeffrey D. Macklis is an American neuroscientist. He is the Max and Anne Wien Professor of Life Sciences in the Department of Stem Cell and Regenerative Biology and Center for Brain Science at Harvard University, Professor of Neurology [Neuroscience] at Harvard Medical School, and on the Executive Committee and a Member of the Principal Faculty of the Neuroscience / Nervous System Diseases Program at the Harvard Stem Cell Institute.