Brain mapping is a set of neuroscience techniques predicated on the mapping of (biological) quantities or properties onto spatial representations of the brain resulting in maps.
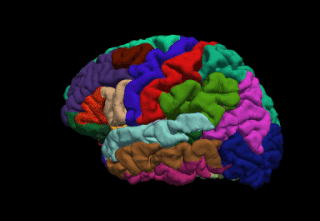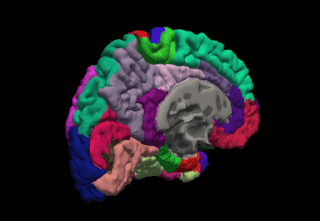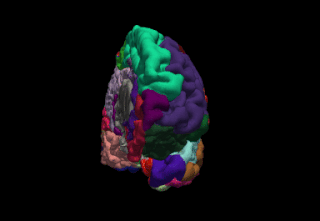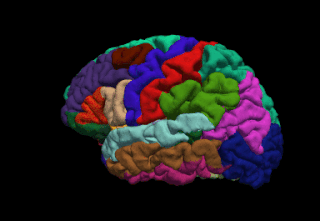
FreeSurfer is a brain imaging software package originally developed by Bruce Fischl, Anders Dale, Martin Sereno, and Doug Greve. Development and maintenance of FreeSurfer is now the primary responsibility of the Laboratory for Computational Neuroimaging at the Athinoula A. Martinos Center for Biomedical Imaging. FreeSurfer contains a set of programs with a common focus of analyzing magnetic resonance imaging (MRI) scans of brain tissue. It is an important tool in functional brain mapping and contains tools to conduct both volume based and surface based analysis. FreeSurfer includes tools for the reconstruction of topologically correct and geometrically accurate models of both the gray/white and pial surfaces, for measuring cortical thickness, surface area and folding, and for computing inter-subject registration based on the pattern of cortical folds.

Talairach coordinates, also known as Talairach space, is a 3-dimensional coordinate system of the human brain, which is used to map the location of brain structures independent from individual differences in the size and overall shape of the brain. It is still common to use Talairach coordinates in functional brain imaging studies and to target transcranial stimulation of brain regions. However, alternative methods such as the MNI Coordinate System have largely replaced Talairach for stereotaxy and other procedures.
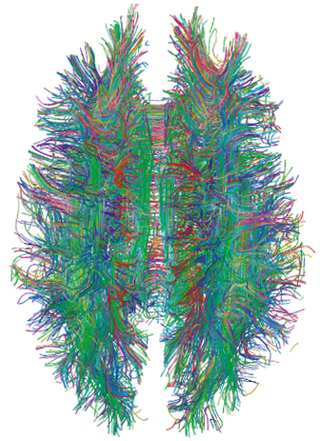
A connectome is a comprehensive map of neural connections in the brain, and may be thought of as its "wiring diagram". An organism's nervous system is made up of neurons which communicate through synapses. A connectome is constructed by tracing the neuron in a nervous system and mapping where neurons are connected through synapses.
Connectomics is the production and study of connectomes: comprehensive maps of connections within an organism's nervous system. More generally, it can be thought of as the study of neuronal wiring diagrams with a focus on how structural connectivity, individual synapses, cellular morphology, and cellular ultrastructure contribute to the make up of a network. The nervous system is a network made of billions of connections and these connections are responsible for our thoughts, emotions, actions, memories, function and dysfunction. Therefore, the study of connectomics aims to advance our understanding of mental health and cognition by understanding how cells in the nervous system are connected and communicate. Because these structures are extremely complex, methods within this field use a high-throughput application of functional and structural neural imaging, most commonly magnetic resonance imaging (MRI), electron microscopy, and histological techniques in order to increase the speed, efficiency, and resolution of these nervous system maps. To date, tens of large scale datasets have been collected spanning the nervous system including the various areas of cortex, cerebellum, the retina, the peripheral nervous system and neuromuscular junctions.

In neuroscience, the default mode network (DMN), also known as the default network, default state network, or anatomically the medial frontoparietal network (M-FPN), is a large-scale brain network primarily composed of the dorsal medial prefrontal cortex, posterior cingulate cortex, precuneus and angular gyrus. It is best known for being active when a person is not focused on the outside world and the brain is at wakeful rest, such as during daydreaming and mind-wandering. It can also be active during detailed thoughts related to external task performance. Other times that the DMN is active include when the individual is thinking about others, thinking about themselves, remembering the past, and planning for the future.
The Human Connectome Project (HCP) is a five-year project sponsored by sixteen components of the National Institutes of Health, split between two consortia of research institutions. The project was launched in July 2009 as the first of three Grand Challenges of the NIH's Blueprint for Neuroscience Research. On September 15, 2010, the NIH announced that it would award two grants: $30 million over five years to a consortium led by Washington University in St. Louis and the University of Minnesota, with strong contributions from University of Oxford (FMRIB) and $8.5 million over three years to a consortium led by Harvard University, Massachusetts General Hospital and the University of California Los Angeles.

Resting state fMRI is a method of functional magnetic resonance imaging (fMRI) that is used in brain mapping to evaluate regional interactions that occur in a resting or task-negative state, when an explicit task is not being performed. A number of resting-state brain networks have been identified, one of which is the default mode network. These brain networks are observed through changes in blood flow in the brain which creates what is referred to as a blood-oxygen-level dependent (BOLD) signal that can be measured using fMRI.
The biological basis of personality it is the collection of brain systems and mechanisms that underlie human personality. Human neurobiology, especially as it relates to complex traits and behaviors, is not well understood, but research into the neuroanatomical and functional underpinnings of personality are an active field of research. Animal models of behavior, molecular biology, and brain imaging techniques have provided some insight into human personality, especially trait theories.
Gitte Moos Knudsen is a Danish translational neurobiologist and clinical neurologist, and Clinical Professor and Chief Physician at the Department of Clinical Medicine, Neurology, Psychiatry and Sensory Sciences, at Copenhagen University Hospital. She graduated from Lyngby Statsskole, just north of Copenhagen, before gaining entrance to medicine, where she received her MD from University of Copenhagen in 1984, and became a Board certified user of radioisotopes in 1986. She sat the FMGEMS exam (US) in 1989. She became Board certified in neurology in 1994 and received her DMSc (Dr.Med.) from University of Copenhagen in 1994. She currently resides in Copenhagen, and is married to Tore Vulpius. She has 3 children.
The following outline is provided as an overview of and topical guide to brain mapping:
Connectograms are graphical representations of connectomics, the field of study dedicated to mapping and interpreting all of the white matter fiber connections in the human brain. These circular graphs based on diffusion MRI data utilize graph theory to demonstrate the white matter connections and cortical characteristics for single structures, single subjects, or populations.
Dynamic functional connectivity (DFC) refers to the observed phenomenon that functional connectivity changes over a short time. Dynamic functional connectivity is a recent expansion on traditional functional connectivity analysis which typically assumes that functional networks are static in time. DFC is related to a variety of different neurological disorders, and has been suggested to be a more accurate representation of functional brain networks. The primary tool for analyzing DFC is fMRI, but DFC has also been observed with several other mediums. DFC is a recent development within the field of functional neuroimaging whose discovery was motivated by the observation of temporal variability in the rising field of steady state connectivity research.
Large-scale brain networks are collections of widespread brain regions showing functional connectivity by statistical analysis of the fMRI BOLD signal or other recording methods such as EEG, PET and MEG. An emerging paradigm in neuroscience is that cognitive tasks are performed not by individual brain regions working in isolation but by networks consisting of several discrete brain regions that are said to be "functionally connected". Functional connectivity networks may be found using algorithms such as cluster analysis, spatial independent component analysis (ICA), seed based, and others. Synchronized brain regions may also be identified using long-range synchronization of the EEG, MEG, or other dynamic brain signals.
Brain–body interactions are patterns of neural activity in the central nervous system to coordinate the activity between the brain and body. The nervous system consists of central and peripheral nervous systems and coordinates the actions of an animal by transmitting signals to and from different parts of its body. The brain and spinal cord are interwoven with the body and interact with other organ systems through the somatic, autonomic and enteric nervous systems. Neural pathways regulate brain–body interactions and allow to sense and control its body and interact with the environment.
Social cognitive neuroscience is the scientific study of the biological processes underpinning social cognition. Specifically, it uses the tools of neuroscience to study "the mental mechanisms that create, frame, regulate, and respond to our experience of the social world". Social cognitive neuroscience uses the epistemological foundations of cognitive neuroscience, and is closely related to social neuroscience. Social cognitive neuroscience employs human neuroimaging, typically using functional magnetic resonance imaging (fMRI). Human brain stimulation techniques such as transcranial magnetic stimulation and transcranial direct-current stimulation are also used. In nonhuman animals, direct electrophysiological recordings and electrical stimulation of single cells and neuronal populations are utilized for investigating lower-level social cognitive processes.
David C. Van Essen is an American neuroscientist specializing in neurobiology and studies the structure, function, development, connectivity and evolution of the cerebral cortex of humans and nonhuman relatives. After over two decades of teaching at the Washington University in St. Louis School of Medicine, he currently serves as an Alumni Endowed Professor of Neuroscience and maintains an active laboratory. Van Essen has held numerous positions, including Editor-in-Chief of the Journal of Neuroscience, Secretary of the Society for Neuroscience, and the President of the Society for Neuroscience from 2006 to 2007. Additionally, Van Essen has received numerous awards for his efforts in education and science, including the Krieg Cortical Discoverer Award from the Cajal Club in 2002, the Peter Raven Lifetime Achievement Award from St. Louis Academy of Science in 2007, and the Second Century Award in 2015 and the Distinguished Educator Award in 2017, both from Washington University School of Medicine.
Network neuroscience is an approach to understanding the structure and function of the human brain through an approach of network science, through the paradigm of graph theory. A network is a connection of many brain regions that interact with each other to give rise to a particular function. Network Neuroscience is a broad field that studies the brain in an integrative way by recording, analyzing, and mapping the brain in various ways. The field studies the brain at multiple scales of analysis to ultimately explain brain systems, behavior, and dysfunction of behavior in psychiatric and neurological diseases. Network neuroscience provides an important theoretical base for understanding neurobiological systems at multiple scales of analysis.

Dimitri Van De Ville is a Swiss and Belgian computer scientist and neuroscientist specialized in dynamical and network aspects of brain activity. He is a professor of bioengineering at EPFL and the head of the Medical Image Processing Laboratory at EPFL's School of Engineering.






