The Allen Mouse and Human Brain Atlases are projects within the Allen Institute for Brain Science which seek to combine genomics with neuroanatomy by creating gene expression maps for the mouse and human brain. They were initiated in September 2003 with a $100 million donation from Paul G. Allen and the first atlas went public in September 2006. As of May 2012, seven brain atlases have been published: Mouse Brain Atlas, Human Brain Atlas, Developing Mouse Brain Atlas, Developing Human Brain Atlas, Mouse Connectivity Atlas, Non-Human Primate Atlas, and Mouse Spinal Cord Atlas. There are also three related projects with data banks: Glioblastoma, Mouse Diversity, and Sleep. It is the hope of the Allen Institute that their findings will help advance various fields of science, especially those surrounding the understanding of neurobiological diseases. The atlases are free and available for public use online.
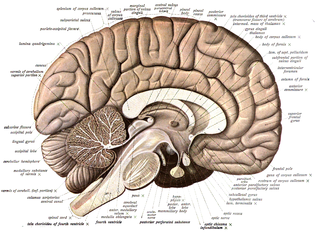
Neuroanatomy is the study of the structure and organization of the nervous system. In contrast to animals with radial symmetry, whose nervous system consists of a distributed network of cells, animals with bilateral symmetry have segregated, defined nervous systems. Their neuroanatomy is therefore better understood. In vertebrates, the nervous system is segregated into the internal structure of the brain and spinal cord and the series of nerves that connect the CNS to the rest of the body. Breaking down and identifying specific parts of the nervous system has been crucial for figuring out how it operates. For example, much of what neuroscientists have learned comes from observing how damage or "lesions" to specific brain areas affects behavior or other neural functions.

A neuroscientist is a scientist who has specialised knowledge in neuroscience, a branch of biology that deals with the physiology, biochemistry, psychology, anatomy and molecular biology of neurons, neural circuits, and glial cells and especially their behavioral, biological, and psychological aspect in health and disease.
Fred "Rusty" Gage is an American geneticist known for his discovery of stem cells in the adult human brain. Gage is a former president (2018–2023) of the Salk Institute for Biological Studies, where he holds the Vi and John Adler Chair for Research on Age-Related Neurodegenerative Disease and works in the Laboratory of Genetics.
Neuroinformatics is the emergent field that combines informatics and neuroscience. Neuroinformatics is related with neuroscience data and information processing by artificial neural networks. There are three main directions where neuroinformatics has to be applied:
The Blue Brain Project is a Swiss brain research initiative that aims to create a digital reconstruction of the mouse brain. The project was founded in May 2005 by the Brain Mind Institute of École Polytechnique Fédérale de Lausanne (EPFL) in Switzerland. Its mission is to use biologically-detailed digital reconstructions and simulations of the mammalian brain to identify the fundamental principles of brain structure and function.
Brain mapping is a set of neuroscience techniques predicated on the mapping of (biological) quantities or properties onto spatial representations of the brain resulting in maps.
The Neuroscience Information Framework is a repository of global neuroscience web resources, including experimental, clinical, and translational neuroscience databases, knowledge bases, atlases, and genetic/genomic resources and provides many authoritative links throughout the neuroscience portal of Wikipedia.
Connectomics is the production and study of connectomes: comprehensive maps of connections within an organism's nervous system. More generally, it can be thought of as the study of neuronal wiring diagrams with a focus on how structural connectivity, individual synapses, cellular morphology, and cellular ultrastructure contribute to the make up of a network. The nervous system is a network made of billions of connections and these connections are responsible for our thoughts, emotions, actions, memories, function and dysfunction. Therefore, the study of connectomics aims to advance our understanding of mental health and cognition by understanding how cells in the nervous system are connected and communicate. Because these structures are extremely complex, methods within this field use a high-throughput application of functional and structural neural imaging, most commonly magnetic resonance imaging (MRI), electron microscopy, and histological techniques in order to increase the speed, efficiency, and resolution of these nervous system maps. To date, tens of large scale datasets have been collected spanning the nervous system including the various areas of cortex, cerebellum, the retina, the peripheral nervous system and neuromuscular junctions.
Nervous system diseases, also known as nervous system or neurological disorders, refers to a small class of medical conditions affecting the nervous system. This category encompasses over 600 different conditions, including genetic disorders, infections, cancer, seizure disorders, conditions with a cardiovascular origin, congenital and developmental disorders, and degenerative disorders.

David J. Anderson is an American neurobiologist. He is a Howard Hughes Medical Institute investigator. His lab is located at the California Institute of Technology, where he currently holds the position of Seymour Benzer Professor of Biology, TianQiao and Chrissy Chen Leadership Chair and Director, TianQiao and Chrissy Chen Institute for Neuroscience. Anderson is a founding adviser of the Allen Institute for Brain Research, a non-profit research institute funded by the late Paul G. Allen, and spearheaded the Institute's early effort to generate a comprehensive map of gene expression in the mouse brain.

Homeobox protein DBX2, also known as developing brain homeobox protein 2, is a protein that in humans is encoded by the DBX2 gene. DBX2, a homeodomain-containing protein, plays an important role in the development of the central nervous system, specifically in the development of the neural tube and brain. The gene DBX2 is located on chromosome 12 and is approximately 36,000 base pairs long. DBX2 is predicted to enable DNA-binding transcription activity as well as being involved in the regulation of transcription by RNA polymerase II.

Neuroscience Research Australia is an independent, not for profit medical research institute based in Sydney, Australia.

Angela Jane Roskams is a neuroscientist at the University of British Columbia (UBC) with a joint appointment in Neurosurgery at the University of Washington. She is professor at the Centre for Brain Health at UBC, and directed the laboratory of neural regeneration and brain repair, before winding down her lab in 2015–16 to become Executive Director of the Allen Institute for Brain Science, and a leader in the Open Science movement. After leading Strategy and Alliances for the Allen institute's multiple branches, she has become an influencer in the fields of neuroinformatics, public-private partnerships, and Open Data Sharing.
The following outline is provided as an overview of and topical guide to brain mapping:

GPATCH11 is a protein that in humans is encoded by the G-patch domain containing protein 11 gene. The gene has four transcript variants encoding two functional protein isoforms and is expressed in most human tissues. The protein has been found to interact with several other proteins, including two from a splicing pathway. In addition, GPATCH11 has orthologs in all taxa of the eukarya domain.

The Allen Institute is a non-profit, bioscience research institute located in Seattle. It was founded by billionaire philanthropist Paul G. Allen in 2003. The Allen Institute conducts large-scale basic science research studying the brain, cells and immune system in effort to accelerate science and disease research. The organization practices open science, in that they make all their data and resources publicly available for researchers to access.
The Human Cell Atlas is a project to describe all cell types in the human body. The initiative was announced by a consortium after its inaugural meeting in London in October 2016, which established the first phase of the project. Aviv Regev and Sarah Teichmann defined the goals of the project at that meeting, which was convened by the Broad Institute, the Wellcome Trust Sanger Institute and Wellcome Trust. Regev and Teichmann lead the project.
Hongkui Zeng is the Director of the Allen Institute for Brain Science in Seattle, where she leads the creation of open-access datasets and tools to accelerate neuroscience discovery. In 2011-2014 Zeng led the team that created the Allen Mouse Brain Connectivity Atlas, which indicates which regions of the mouse brain are connected to which other regions. Since then, she has led the creation of atlases of neuronal cell types in the brain of humans and mice.










