Related Research Articles
A cell type is a classification used to identify cells that share morphological or phenotypical features. A multicellular organism may contain cells of a number of widely differing and specialized cell types, such as muscle cells and skin cells, that differ both in appearance and function yet have identical genomic sequences. Cells may have the same genotype, but belong to different cell types due to the differential regulation of the genes they contain. Classification of a specific cell type is often done through the use of microscopy. Recent developments in single cell RNA sequencing facilitated classification of cell types based on shared gene expression patterns. This has led to the discovery of many new cell types in e.g. mouse cortex, hippocampus, dorsal root ganglion and spinal cord.
The Allen Mouse and Human Brain Atlases are projects within the Allen Institute for Brain Science which seek to combine genomics with neuroanatomy by creating gene expression maps for the mouse and human brain. They were initiated in September 2003 with a $100 million donation from Paul G. Allen and the first atlas went public in September 2006. As of May 2012, seven brain atlases have been published: Mouse Brain Atlas, Human Brain Atlas, Developing Mouse Brain Atlas, Developing Human Brain Atlas, Mouse Connectivity Atlas, Non-Human Primate Atlas, and Mouse Spinal Cord Atlas. There are also three related projects with data banks: Glioblastoma, Mouse Diversity, and Sleep. It is the hope of the Allen Institute that their findings will help advance various fields of science, especially those surrounding the understanding of neurobiological diseases. The atlases are free and available for public use online.
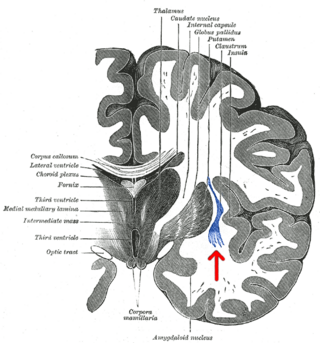
The claustrum is a thin sheet of neurons and supporting glial cells, that connects to the cerebral cortex and subcortical regions including the amygdala, hippocampus and thalamus of the brain. It is located between the insular cortex laterally and the putamen medially, encased by the extreme and external capsules respectively. Blood to the claustrum is supplied by the middle cerebral artery. It is considered to be the most densely connected structure in the brain, and thus hypothesized to allow for the integration of various cortical inputs such as vision, sound and touch, into one experience. Other hypotheses suggest that the claustrum plays a role in salience processing, to direct attention towards the most behaviorally relevant stimuli amongst the background noise. The claustrum is difficult to study given the limited number of individuals with claustral lesions and the poor resolution of neuroimaging.
The Blue Brain Project is a Swiss brain research initiative that aims to create a digital reconstruction of the mouse brain. The project was founded in May 2005 by the Brain Mind Institute of École Polytechnique Fédérale de Lausanne (EPFL) in Switzerland. Its mission is to use biologically-detailed digital reconstructions and simulations of the mammalian brain to identify the fundamental principles of brain structure and function.
Brain mapping is a set of neuroscience techniques predicated on the mapping of (biological) quantities or properties onto spatial representations of the brain resulting in maps.

Brainbow is a process by which individual neurons in the brain can be distinguished from neighboring neurons using fluorescent proteins. By randomly expressing different ratios of red, green, and blue derivatives of green fluorescent protein in individual neurons, it is possible to flag each neuron with a distinctive color. This process has been a major contribution to the field of neural connectomics.

Probable G-protein coupled receptor 37 is a protein that in humans is encoded by the GPR37 gene. GPR37 is primarily found in the central nervous system (CNS), with significant expression observed in various CNS regions including the amygdala, basal ganglia, substantia nigra, hippocampus, frontal cortex, and hypothalamus, particularly noteworthy is its exceptionally elevated expression in the spinal cord.
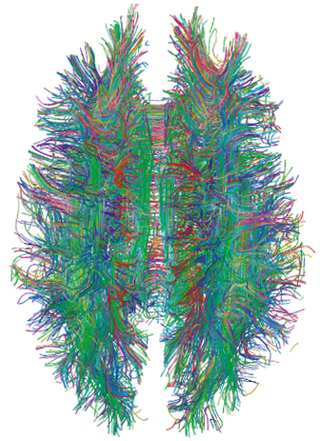
A connectome is a comprehensive map of neural connections in the brain, and may be thought of as its "wiring diagram". An organism's nervous system is made up of neurons which communicate through synapses. A connectome is constructed by tracing the neuron in a nervous system and mapping where neurons are connected through synapses.

The Allen Institute for Brain Science is a division of the Allen Institute, based in Seattle, Washington, that focuses on bioscience research. Founded in 2003, it is dedicated to accelerating the understanding of how the human brain works. With the intent of catalyzing brain research in different areas, the Allen Institute provides free data and tools to scientists.
Connectomics is the production and study of connectomes: comprehensive maps of connections within an organism's nervous system. More generally, it can be thought of as the study of neuronal wiring diagrams with a focus on how structural connectivity, individual synapses, cellular morphology, and cellular ultrastructure contribute to the make up of a network. The nervous system is a network made of billions of connections and these connections are responsible for our thoughts, emotions, actions, memories, function and dysfunction. Therefore, the study of connectomics aims to advance our understanding of mental health and cognition by understanding how cells in the nervous system are connected and communicate. Because these structures are extremely complex, methods within this field use a high-throughput application of functional and structural neural imaging, most commonly magnetic resonance imaging (MRI), electron microscopy, and histological techniques in order to increase the speed, efficiency, and resolution of these nervous system maps. To date, tens of large scale datasets have been collected spanning the nervous system including the various areas of cortex, cerebellum, the retina, the peripheral nervous system and neuromuscular junctions.

GCaMP is a genetically encoded calcium indicator (GECI) initially developed in 2001 by Junichi Nakai. It is a synthetic fusion of green fluorescent protein (GFP), calmodulin (CaM), and M13, a peptide sequence from myosin light-chain kinase. When bound to Ca2+, GCaMP fluoresces green with a peak excitation wavelength of 480 nm and a peak emission wavelength of 510 nm. It is used in biological research to measure intracellular Ca2+ levels both in vitro and in vivo using virally transfected or transgenic cell and animal lines. The genetic sequence encoding GCaMP can be inserted under the control of promoters exclusive to certain cell types, allowing for cell-type specific expression of GCaMP. Since Ca2+ is a second messenger that contributes to many cellular mechanisms and signaling pathways, GCaMP allows researchers to quantify the activity of Ca2+-based mechanisms and study the role of Ca2+ ions in biological processes of interest.
The White House BRAIN Initiative is a collaborative, public-private research initiative announced by the Obama administration on April 2, 2013, with the goal of supporting the development and application of innovative technologies that can create a dynamic understanding of brain function.
The following outline is provided as an overview of and topical guide to brain mapping:

Douglas G. McMahon is a professor of Biological Sciences and Pharmacology at Vanderbilt University. McMahon has contributed several important discoveries to the field of chronobiology and vision. His research focuses on connecting the anatomical location in the brain to specific behaviors. As a graduate student under Gene Block, McMahon identified that the basal retinal neurons (BRNs) of the molluscan eye exhibited circadian rhythms in spike frequency and membrane potential, indicating they are the clock neurons. He became the 1986 winner of the Society for Neuroscience's Donald B. Lindsley Prize in Behavioral Neuroscience for his work. Later, he moved on to investigate visual, circadian, and serotonergic mechanisms of neuroplasticity. In addition, he helped find that constant light can desynchronize the circadian cells in the suprachiasmatic nucleus (SCN). He has always been interested in the underlying causes of behavior and examining the long term changes in behavior and physiology in the neurological modular system. McMahon helped identifying a retrograde neurotransmission system in the retina involving the melanopsin containing ganglion cells and the retinal dopaminergic amacrine neurons.

The Allen Institute is a non-profit, bioscience research institute located in Seattle. It was founded by billionaire philanthropist Paul G. Allen in 2003. The Allen Institute conducts large-scale basic science research studying the brain, cells and immune system in effort to accelerate science and disease research. The organization practices open science, in that they make all their data and resources publicly available for researchers to access.

The mouse brain refers to the brain of Mus musculus. Various brain atlases exist.
Richard Palmiter is a cellular biologist. He was born in Poughkeepsie, NY, and later went on to earn a BA in Zoology from Duke University and a PhD in Biological Sciences from Stanford University. He is employed with the University of Washington where he is a professor of biochemistry and genome sciences. His current research involves developing a deeper understanding of Parkinson's disease. His most notable research is a collaboration with Dr. Ralph Brinster where they injected purified DNA into a single-cell mouse embryo, showing transmission of the genetic material to subsequent generations for the first time.
Susan M. Dymecki is an American geneticist and neuroscientist and director of the Biological and Biomedical Sciences PhD Program at Harvard University. Dymecki is also a professor in the Department of Genetics and the principal investigator of the Dymecki Lab at Harvard. Her lab characterizes the development and function of unique populations of serotonergic neurons in the mouse brain. To enable this functional dissection, Dymecki has pioneered several transgenic tools for probing neural circuit development and function. Dymecki also competed internationally as an ice dancer, placing 7th in the 1980 U.S. Figure Skating Championships.
Fan Wang is a neuroscientist and professor in the MIT Department of Brain and Cognitive Sciences. She is an investigator at the McGovern Institute for Brain Research. Wang is known for her work identifying neural circuits underlying touch, pain, and anesthesia; and the development of a technique for capturing activated neuronal ensembles (CANE) to label and manipulate neurons activated by stimuli or behavioral paradigms.
References
- ↑ "Hongkui Zeng". alleninstitute.org. Retrieved 2021-05-10.
- 1 2 Begley, Sharon (2014-04-07). "Researchers report that they have created a diagram of a mouse's brain". Washington Post. ISSN 0190-8286 . Retrieved 2021-05-10.
- 1 2 Dance, Amber. "A Massive Global Effort Maps How the Brain Is Wired". Scientific American. Retrieved 2021-05-10.
- 1 2 "Neuroscientists draw up a 'parts list' covering 133 different types of brain cells". GeekWire. 2018-11-01. Retrieved 2021-05-10.
- 1 2 "Revealed: New Cortical Neuron Types". The Scientist Magazine®. Retrieved 2021-05-10.
- 1 2 "Overview :: Allen Brain Atlas: Cell Types". celltypes.brain-map.org. Retrieved 2021-05-10.
- 1 2 "Transcriptional Landscape of the Brain - brain-map.org". portal.brain-map.org. Retrieved 2021-05-10.
- ↑ Oh, Seung Wook; Harris, Julie A.; Ng, Lydia; Winslow, Brent; Cain, Nicholas; Mihalas, Stefan; Wang, Quanxin; Lau, Chris; Kuan, Leonard; Henry, Alex M.; Mortrud, Marty T. (April 2014). "A mesoscale connectome of the mouse brain". Nature. 508 (7495): 207–214. Bibcode:2014Natur.508..207O. doi:10.1038/nature13186. ISSN 1476-4687. PMC 5102064 . PMID 24695228.
- ↑ Marx, Vivien (January 2021). "Method of the Year: spatially resolved transcriptomics". Nature Methods. 18 (1): 9–14. doi: 10.1038/s41592-020-01033-y . ISSN 1548-7105. PMID 33408395.
- ↑ "Pradel Research Award". National Academy of Sciences.
- ↑ "National Academy of Sciences Member Directory". National Academy of Sciences.