In chemistry, biochemistry, and pharmacology, a dissociation constant (KD) is a specific type of equilibrium constant that measures the propensity of a larger object to separate (dissociate) reversibly into smaller components, as when a complex falls apart into its component molecules, or when a salt splits up into its component ions. The dissociation constant is the inverse of the association constant. In the special case of salts, the dissociation constant can also be called an ionization constant. For a general reaction:

Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity.

In the fields of biochemistry and pharmacology an allosteric regulator is a substance that binds to a site on an enzyme or receptor distinct from the active site, resulting in a conformational change that alters the protein's activity, either enhancing or inhibiting its function. In contrast, substances that bind directly to an enzyme's active site or the binding site of the endogenous ligand of a receptor are called orthosteric regulators or modulators.
In molecular biology and pharmacology, a small molecule or micromolecule is a low molecular weight organic compound that may regulate a biological process, with a size on the order of 1 nm. Many drugs are small molecules; the terms are equivalent in the literature. Larger structures such as nucleic acids and proteins, and many polysaccharides are not small molecules, although their constituent monomers are often considered small molecules. Small molecules may be used as research tools to probe biological function as well as leads in the development of new therapeutic agents. Some can inhibit a specific function of a protein or disrupt protein–protein interactions.

In biochemistry and molecular biology, a binding site is a region on a macromolecule such as a protein that binds to another molecule with specificity. The binding partner of the macromolecule is often referred to as a ligand. Ligands may include other proteins, enzyme substrates, second messengers, hormones, or allosteric modulators. The binding event is often, but not always, accompanied by a conformational change that alters the protein's function. Binding to protein binding sites is most often reversible, but can also be covalent reversible or irreversible.
A hormone receptor is a receptor molecule that binds to a specific hormone. Hormone receptors are a wide family of proteins made up of receptors for thyroid and steroid hormones, retinoids and Vitamin D, and a variety of other receptors for various ligands, such as fatty acids and prostaglandins. Hormone receptors are of mainly two classes. Receptors for peptide hormones tend to be cell surface receptors built into the plasma membrane of cells and are thus referred to as trans membrane receptors. An example of this is Actrapid. Receptors for steroid hormones are usually found within the protoplasm and are referred to as intracellular or nuclear receptors, such as testosterone. Upon hormone binding, the receptor can initiate multiple signaling pathways, which ultimately leads to changes in the behavior of the target cells.
Affinity chromatography is a method of separating a biomolecule from a mixture, based on a highly specific macromolecular binding interaction between the biomolecule and another substance. The specific type of binding interaction depends on the biomolecule of interest; antigen and antibody, enzyme and substrate, receptor and ligand, or protein and nucleic acid binding interactions are frequently exploited for isolation of various biomolecules. Affinity chromatography is useful for its high selectivity and resolution of separation, compared to other chromatographic methods.

Ion chromatography is a form of chromatography that separates ions and ionizable polar molecules based on their affinity to the ion exchanger. It works on almost any kind of charged molecule—including small inorganic anions, large proteins, small nucleotides, and amino acids. However, ion chromatography must be done in conditions that are one pH unit away from the isoelectric point of a protein.
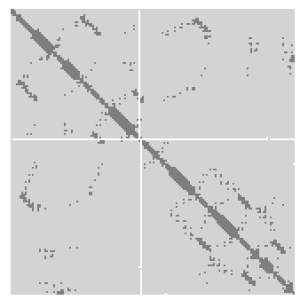
A protein contact map represents the distance between all possible amino acid residue pairs of a three-dimensional protein structure using a binary two-dimensional matrix. For two residues and , the element of the matrix is 1 if the two residues are closer than a predetermined threshold, and 0 otherwise. Various contact definitions have been proposed: The distance between the Cα-Cα atom with threshold 6-12 Å; distance between Cβ-Cβ atoms with threshold 6-12 Å ; and distance between the side-chain centers of mass.

The EF hand is a helix–loop–helix structural domain or motif found in a large family of calcium-binding proteins.

PK-11195 is an isoquinoline carboxamide which binds selectively to the peripheral benzodiazepine receptor (PBR). It is one of the most commonly used PBR ligands due to its high affinity for the PBR in all species, although it is starting to be replaced by newer and more selective ligands.

The lipocalins are a family of proteins which transport small hydrophobic molecules such as steroids, bilins, retinoids, and lipids, and most lipocalins are also able to bind to complexed iron as well as heme. They share limited regions of sequence homology and a common tertiary structure architecture. This is an eight stranded antiparallel beta barrel with a repeated + 1 topology enclosing an internal ligand binding site.

Bilins, bilanes or bile pigments are biological pigments formed in many organisms as a metabolic product of certain porphyrins. Bilin was named as a bile pigment of mammals, but can also be found in lower vertebrates, invertebrates, as well as red algae, green plants and cyanobacteria. Bilins can range in color from red, orange, yellow or brown to blue or green.

Odorant-binding protein 2a is a protein that in humans is encoded by the OBP2A gene.
Antibody mimetics are organic compounds that, like antibodies, can specifically bind antigens, but that are not structurally related to antibodies. They are usually artificial peptides or proteins with a molar mass of about 3 to 20 kDa.

Affitins are artificial proteins with the ability to selectively bind antigens. They are structurally derived from the DNA binding protein Sac7d, found in Sulfolobus acidocaldarius, a microorganism belonging to the archaeal domain. By randomizing the amino acids on the binding surface of Sac7d and subjecting the resulting protein library to rounds of ribosome display, the affinity can be directed towards various targets, such as peptides, proteins, viruses, and bacteria.
Affilins are artificial proteins designed to selectively bind antigens. Affilin proteins are structurally derived from human ubiquitin. Affilin proteins are constructed by modification of surface-exposed amino acids of these proteins and isolated by display techniques such as phage display and screening. They resemble antibodies in their affinity and specificity to antigens but not in structure, which makes them a type of antibody mimetic. Affilin was developed by Scil Proteins GmbH as potential new biopharmaceutical drugs, diagnostics and affinity ligands.
A ligand binding assay (LBA) is an assay, or an analytic procedure, which relies on the binding of ligand molecules to receptors, antibodies or other macromolecules. A detection method is used to determine the presence and amount of the ligand-receptor complexes formed, and this is usually determined electrochemically or through a fluorescence detection method. This type of analytic test can be used to test for the presence of target molecules in a sample that are known to bind to the receptor.
Molecular Operating Environment (MOE) is a drug discovery software platform that integrates visualization, modeling and simulations, as well as methodology development, in one package. MOE scientific applications are used by biologists, medicinal chemists and computational chemists in pharmaceutical, biotechnology and academic research. MOE runs on Windows, Linux, Unix, and macOS. Main application areas in MOE include structure-based design, fragment-based design, ligand-based design, pharmacophore discovery, medicinal chemistry applications, biologics applications, structural biology and bioinformatics, protein and antibody modeling, molecular modeling and simulations, virtual screening, cheminformatics & QSAR. The Scientific Vector Language (SVL) is the built-in command, scripting and application development language of MOE.

Optimer ligands are short synthetic oligonucleotide molecules composed of DNA or RNA that bind to a specific target molecule. They are engineered to bind their target molecules with affinity typically in the low nanomolar range. Optimers can be used as antibody mimetics in a range of applications, and have been optimized to increase their stability, reduce their molecular weight, and offer increased scalability and consistency in manufacture compared to standard aptamer molecules.














