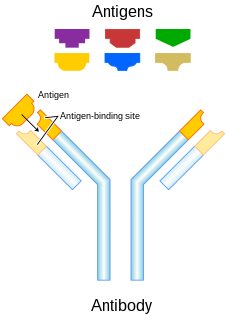
In immunology, an antigen (Ag) is a molecule or molecular structure, such as may be present at the outside of a pathogen, that can be bound by an antigen-specific antibody or B cell antigen receptor. The presence of antigens in the body normally triggers an immune response. The Ag abbreviation stands for an antibody generator.

An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein produced mainly by plasma cells that is used by the immune system to neutralize pathogens such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the pathogen, called an antigen, via the fragment antigen-binding (Fab) variable region. Each tip of the "Y" of an antibody contains a paratope that is specific for one particular epitope on an antigen, allowing these two structures to bind together with precision. Using this binding mechanism, an antibody can tag a microbe or an infected cell for attack by other parts of the immune system, or can neutralize its target directly. Depending on the antigen, the binding may impede the biological process causing the disease or may activate macrophages to destroy the foreign substance. The ability of an antibody to communicate with the other components of the immune system is mediated via its Fc region, which contains a conserved glycosylation site involved in these interactions. The production of antibodies is the main function of the humoral immune system.
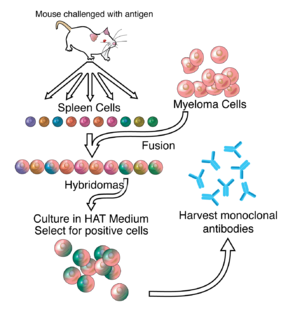
A monoclonal antibody is an antibody made by cloning a unique white blood cell. All subsequent antibodies derived this way trace back to a unique parent cell.

Biochemistry is the study of the chemical processes in living organisms. It deals with the structure and function of cellular components such as proteins, carbohydrates, lipids, nucleic acids and other biomolecules.

A peptidomimetic is a small protein-like chain designed to mimic a peptide. They typically arise either from modification of an existing peptide, or by designing similar systems that mimic peptides, such as peptoids and β-peptides. Irrespective of the approach, the altered chemical structure is designed to advantageously adjust the molecular properties such as stability or biological activity. This can have a role in the development of drug-like compounds from existing peptides. These modifications involve changes to the peptide that will not occur naturally. Based on their similarity with the precursor peptide, peptidomimetics can be grouped into four classes where A features the most and D the least similarities. Classes A and B involve peptide-like scaffolds, while classes C and D include small molecules.

Phage display is a laboratory technique for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages to connect proteins with the genetic information that encodes them. In this technique, a gene encoding a protein of interest is inserted into a phage coat protein gene, causing the phage to "display" the protein on its outside while containing the gene for the protein on its inside, resulting in a connection between genotype and phenotype. These displaying phages can then be screened against other proteins, peptides or DNA sequences, in order to detect interaction between the displayed protein and those other molecules. In this way, large libraries of proteins can be screened and amplified in a process called in vitro selection, which is analogous to natural selection.

A single-domain antibody (sdAb), also known as a nanobody, is an antibody fragment consisting of a single monomeric variable antibody domain. Like a whole antibody, it is able to bind selectively to a specific antigen. With a molecular weight of only 12–15 kDa, single-domain antibodies are much smaller than common antibodies which are composed of two heavy protein chains and two light chains, and even smaller than Fab fragments and single-chain variable fragments.
Humanized antibodies are antibodies from non-human species whose protein sequences have been modified to increase their similarity to antibody variants produced naturally in humans. The process of "humanization" is usually applied to monoclonal antibodies developed for administration to humans. Humanization can be necessary when the process of developing a specific antibody involves generation in a non-human immune system. The protein sequences of antibodies produced in this way are partially distinct from homologous antibodies occurring naturally in humans, and are therefore potentially immunogenic when administered to human patients. There are other types of antibodies developed. The International Nonproprietary Names of humanized antibodies end in -zumab, as in omalizumab.

The immunoglobulin superfamily (IgSF) is a large protein superfamily of cell surface and soluble proteins that are involved in the recognition, binding, or adhesion processes of cells. Molecules are categorized as members of this superfamily based on shared structural features with immunoglobulins ; they all possess a domain known as an immunoglobulin domain or fold. Members of the IgSF include cell surface antigen receptors, co-receptors and co-stimulatory molecules of the immune system, molecules involved in antigen presentation to lymphocytes, cell adhesion molecules, certain cytokine receptors and intracellular muscle proteins. They are commonly associated with roles in the immune system. Otherwise, the sperm-specific protein IZUMO1, a member of the immunoglobulin superfamily, has also been identified as the only sperm membrane protein essential for sperm-egg fusion.

mRNA display is a display technique used for in vitro protein, and/or peptide evolution to create molecules that can bind to a desired target. The process results in translated peptides or proteins that are associated with their mRNA progenitor via a puromycin linkage. The complex then binds to an immobilized target in a selection step. The mRNA-protein fusions that bind well are then reverse transcribed to cDNA and their sequence amplified via a polymerase chain reaction. The result is a nucleotide sequence that encodes a peptide with high affinity for the molecule of interest.

In molecular biology, a framework region is a subdivision of the variable region (Fab) of the antibody. The variable region is composed of seven amino acid regions, four of which are framework regions and three of which are hypervariable regions. The framework region makes up about 85% of the variable region. Located on the tips of the Y-shaped molecule, the framework regions are responsible for acting as a scaffold for the complementarity determining regions (CDR), also referred to as hypervariable regions, of the Fab. These CDRs are in direct contact with the antigen and are involved in binding antigen, while the framework regions support the binding of the CDR to the antigen and aid in maintaining the overall structure of the four variable domains on the antibody. To increase its stability, the framework region has less variability in its amino acid sequences compared to the CDR.
Affibody molecules are small, robust proteins engineered to bind to a large number of target proteins or peptides with high affinity, imitating monoclonal antibodies, and are therefore a member of the family of antibody mimetics. Affibody molecules are used in biochemical research and are being developed as potential new biopharmaceutical drugs. These molecules can be used for molecular recognition in diagnostic and therapeutic applications.
Antibody mimetics are organic compounds that, like antibodies, can specifically bind antigens, but that are not structurally related to antibodies. They are usually artificial peptides or proteins with a molar mass of about 3 to 20 kDa.

Monobodies are synthetic binding proteins constructed using a fibronectin type III domain (FN3) as a molecular scaffold. Specifically, this class of binding proteins are built upon a diversified library of the 10th FN3 domain of human fibronectin. Monobodies are a simple and robust alternative to antibodies for creating target-binding proteins. The hybrid term monobody was coined in 1998 by the Koide group who published the first paper demonstrating the monobody concept using the tenth FN3 domain of human fibronectin.
DARPins are genetically engineered antibody mimetic proteins typically exhibiting highly specific and high-affinity target protein binding. They are derived from natural ankyrin proteins, one of the most common classes of binding proteins in nature, which are responsible for diverse functions such as cell signaling, regulation and structural integrity of the cell. DARPins consist of at least three, repeat motifs proteins, and usually consist of four or five. Their molecular mass is about 14 or 18 kDa (kilodaltons) for four- or five-repeat DARPins, respectively.
Avimers are artificial proteins that are able to specifically bind to certain antigens via multiple binding sites. Since they are not structurally related to antibodies, they are classified as a type of antibody mimetic. Avimers have been developed by the biotechnology company Avidia, now part of Amgen, as potential new pharmaceutical drugs.
Synthetic antibodies are affinity reagents generated entirely in vitro, thus completely eliminating animals from the production process. Synthetic antibodies include recombinant antibodies, nucleic acid aptamers and non-immunoglobulin protein scaffolds. As a consequence of their in vitro manufacturing method the antigen recognition site of synthetic antibodies can be engineered to any desired target and may extend beyond the typical immune repertoire offered by natural antibodies. Synthetic antibodies are being developed for use in research, diagnostic and therapeutic applications. Synthetic antibodies can be used in all applications where traditional monoclonal or polyclonal antibodies are used and offer many inherent advantages over animal-derived antibodies, including comparatively low production costs, reagent reproducibility and increased affinity, specificity and stability across a range of experimental conditions.
Affimer molecules are small proteins that bind to target molecules with similar specificity and affinity to that of antibodies. These engineered non-antibody binding proteins are designed to mimic the molecular recognition characteristics of monoclonal antibodies in different applications. In addition, these affinity reagents have been optimized to increase their stability, make them tolerant to a range of temperatures and pH, reduce their size, and to increase their expression in E.coli and mammalian cells.
The tailspike protein (P22TSP) of Enterobacteria phage P22 mediates the recognition and adhesion between the bacteriophage and the surface of Salmonella enterica cells. It is anchored within the viral coat and recognizes the O-antigen portion of the lipopolysaccharide (LPS) on the outer-membrane of Gram-negative bacteria. It possesses endoglycanase activity, serving to shorten the length of the O-antigen during infection.
In the medical field of immunology, nanoCLAMP affinity reagents are recombinant 15 kD antibody mimetic proteins selected for tight, selective and gently reversible binding to target molecules. The nanoCLAMP scaffold is based on an IgG-like, thermostable carbohydrate binding module family 32 (CBM32) from a Clostridium perfringens hyaluronidase. The shape of nanoCLAMPs approximates a cylinder of approximately 4 nm in length and 2.5 nm in diameter, roughly the same size as a nanobody. nanoCLAMPs to specific targets are generated by varying the amino acid sequences and sometimes the length of three solvent exposed, adjacent loops that connect the beta strands making up the beta-sandwich fold, conferring binding affinity and specificity for the target.















